RBP Type
Canonical_RBPs
Diseases
Drug
N.A.
Main interacting RNAs
N.A.
Moonlighting functions
E3 ligase
Localizations
N.A.
BulkPerturb-seq
Ensembl ID ENSG00000101871 Gene ID
4281 Accession
7095
Symbol
MID1
Alias
OS;FXY;OSX;GBBB;OGS1;XPRF;BBBG1;GBBB1;MIDIN;RNF59;ZNFXY;TRIM18
Full Name
midline 1
Status
Confidence
Length
388345 bases
Strand
Minus strand
Position
X : 10445310 - 10833654
RNA binding domain
SPRY
Summary
The protein encoded by this gene is a member of the tripartite motif (TRIM) family, also known as the 'RING-B box-coiled coil' (RBCC) subgroup of RING finger proteins. The TRIM motif includes three zinc-binding domains, a RING, a B-box type 1 and a B-box type 2, and a coiled-coil region. This protein forms homodimers which associate with microtubules in the cytoplasm. The protein is likely involved in the formation of multiprotein structures acting as anchor points to microtubules. Mutations in this gene have been associated with the X-linked form of Opitz syndrome, which is characterized by midline abnormalities such as cleft lip, laryngeal cleft, heart defects, hypospadias, and agenesis of the corpus callosum. This gene was also the first example of a gene subject to X inactivation in human while escaping it in mouse. Alternative promoter use, alternative splicing and alternative polyadenylation result in multiple transcript variants that have different tissue specificities. [provided by RefSeq, Dec 2016]
RNA binding domains (RBDs)
Protein
Domain
Pfam ID
E-value
Domain number
ENSP00000502171
SPRY
PF00622 1.4e-17
1
ENSP00000508663
SPRY
PF00622 4e-17
1
ENSP00000509730
SPRY
PF00622 4e-17
1
ENSP00000509925
SPRY
PF00622 4e-17
1
ENSP00000312678
SPRY
PF00622 4.3e-17
1
ENSP00000370156
SPRY
PF00622 4.3e-17
1
ENSP00000370157
SPRY
PF00622 4.3e-17
1
ENSP00000370162
SPRY
PF00622 4.3e-17
1
ENSP00000370164
SPRY
PF00622 4.3e-17
1
ENSP00000414521
SPRY
PF00622 4.3e-17
1
ENSP00000501707
SPRY
PF00622 4.7e-17
1
RNA binding proteomes (RBPomes)
Pubmed ID
Full Name
Cell
Author
Time
Doi
Literatures on RNA binding capacity
Pubmed ID
Title
Author
Time
Journal
29062069 Resveratrol induces dephosphorylation of Tau by interfering with the MID1-PP2A complex.
Susann Schweiger
2017-10-23
Scientific reports
27774050 Regulation of mRNA Translation by MID1: A Common Mechanism of Expanded CAG Repeat RNAs.
Nadine Griesche
2016-01-01
Frontiers in cellular neuroscience
18949047 Active transport of the ubiquitin ligase MID1 along the microtubules is regulated by protein phosphatase 2A.
Beatriz Aranda-Orgillés
2008-01-01
PloS one
32283114 The MID1 gene product in physiology and disease.
Rossella Baldini
2020-07-15
Gene
37062072 CAG-Repeat RNA Hairpin Folding and Recruitment to Nuclear Speckles with a Pivotal Role of ATP as a Cosolute.
Alexander Hautke
2023-05-03
Journal of the American Chemical Society
24913494 A hormone-dependent feedback-loop controls androgen receptor levels by limiting MID1, a novel translation enhancer and promoter of oncogenic signaling.
Andrea Köhler
2014-06-09
Molecular cancer
27625393 Identifying proteins that bind to specific RNAs - focus on simple repeat expansion diseases.
Magdalena Jazurek
2016-11-02
Nucleic acids research
18005432 Alternative polyadenylation signals and promoters act in concert to control tissue-specific expression of the Opitz Syndrome gene MID1.
Jennifer Winter
2007-11-15
BMC molecular biology
18172692 The Opitz syndrome gene product MID1 assembles a microtubule-associated ribonucleoprotein complex.
Beatriz Aranda-Orgillés
2008-03-01
Human genetics
Name
Transcript ID
bp
Protein
Translation ID
MID1-208
ENST00000453318
6393
667aa
ENSP00000414521
MID1-202
ENST00000380779
6044
667aa
ENSP00000370156
MID1-203
ENST00000380780
6195
667aa
ENSP00000370157
MID1-201
ENST00000317552
6168
667aa
ENSP00000312678
MID1-207
ENST00000413894
6095
667aa
ENSP00000391154
MID1-205
ENST00000380785
6092
667aa
ENSP00000370162
MID1-206
ENST00000380787
5973
667aa
ENSP00000370164
MID1-204
ENST00000380782
3352
552aa
ENSP00000370159
MID1-222
ENST00000691943
3436
631aa
ENSP00000508663
MID1-220
ENST00000690004
3473
629aa
ENSP00000509730
MID1-218
ENST00000689773
3452
629aa
ENSP00000509925
MID1-213
ENST00000675073
2369
718aa
ENSP00000501707
MID1-212
ENST00000674917
1365
371aa
ENSP00000502171
MID1-209
ENST00000479925
858
No protein
-
MID1-216
ENST00000687008
2439
489aa
ENSP00000508734
MID1-221
ENST00000691913
1141
No protein
-
MID1-211
ENST00000616003
1927
540aa
ENSP00000484712
MID1-214
ENST00000675358
557
48aa
ENSP00000502220
MID1-210
ENST00000610939
1839
No protein
-
MID1-215
ENST00000686012
2250
No protein
-
MID1-217
ENST00000689180
1248
No protein
-
MID1-223
ENST00000693721
1992
No protein
-
MID1-219
ENST00000689988
1056
No protein
-
MID1-224
ENST00000707197
1364
No protein
-
MID1-225
ENST00000707198
1305
No protein
-
Pathway ID
Pathway Name
Source
ensgID
Trait
pValue
Pubmed ID
3576 Cholesterol, HDL 2.84798923929218E-7 17903299 6264 Erythrocyte Indices 5.6512429342198E-7 17903294 9781 Echocardiography 2.38083448696652E-6 17903301 105657 Neuropsychological Tests 1E-7 19734545
ensgID SNP Chromosome Position
Trait PubmedID Or or BEAT
EFO ID
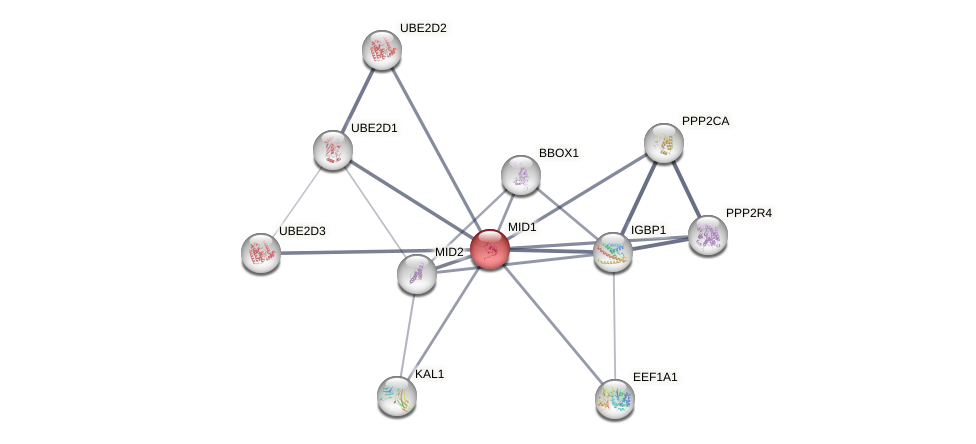
Protein-Protein Interaction (PPI)
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Ensembl ID
Source
Target
Species
Protein ID
Perc_pos
Perc_id
Species
Protein ID
Perc_pos
Perc_id
Go ID
Go term
No. evidence
Entries
Species
Category
GO:0000226 involved_in microtubule cytoskeleton organization 1 TAS Homo_sapiens(9606) Process GO:0005515 enables protein binding 1 IPI Homo_sapiens(9606) Function GO:0005737 is_active_in cytoplasm 1 IBA Homo_sapiens(9606) Component GO:0005794 located_in Golgi apparatus 1 IDA Homo_sapiens(9606) Component GO:0005819 located_in spindle 1 IEA Homo_sapiens(9606) Component GO:0005829 located_in cytosol 2 IDA,TAS Homo_sapiens(9606) Component GO:0005874 located_in microtubule 1 IDA Homo_sapiens(9606) Component GO:0005875 part_of microtubule associated complex 1 TAS Homo_sapiens(9606) Component GO:0005881 colocalizes_with cytoplasmic microtubule 1 IDA Homo_sapiens(9606) Component GO:0007026 involved_in negative regulation of microtubule depolymerization 1 IEA Homo_sapiens(9606) Process GO:0007389 involved_in pattern specification process 1 TAS Homo_sapiens(9606) Process GO:0008017 enables microtubule binding 1 IMP Homo_sapiens(9606) Function GO:0008270 enables zinc ion binding 1 IEA Homo_sapiens(9606) Function GO:0016740 enables transferase activity 1 IEA Homo_sapiens(9606) Function GO:0019899 enables enzyme binding 1 IPI Homo_sapiens(9606) Function GO:0031625 enables ubiquitin protein ligase binding 1 IPI Homo_sapiens(9606) Function GO:0032874 involved_in positive regulation of stress-activated MAPK cascade 1 IMP Homo_sapiens(9606) Process GO:0034451 located_in centriolar satellite 1 IDA Homo_sapiens(9606) Component GO:0035372 involved_in protein localization to microtubule 1 IMP Homo_sapiens(9606) Process GO:0042802 enables identical protein binding 1 IPI Homo_sapiens(9606) Function GO:0042803 enables protein homodimerization activity 1 IDA Homo_sapiens(9606) Function GO:0051219 enables phosphoprotein binding 1 IPI Homo_sapiens(9606) Function GO:0070507 involved_in regulation of microtubule cytoskeleton organization 1 IBA Homo_sapiens(9606) Process
Copyright © 2023, Bioinformatics Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, China. All Rights Reserved.