Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000100030 | Gene ID | 5594 | Accession | 6871 |
| Symbol | MAPK1 | Alias | ERK;p38;p40;p41;ERK2;ERT1;NS13;ERK-2;MAPK2;PRKM1;PRKM2;P42MAPK;p41mapk;p42-MAPK | Full Name | mitogen-activated protein kinase 1 |
| Status | Confidence | Length | 108024 bases | Strand | Minus strand |
| Position | 22 : 21759657 - 21867680 | RNA binding domain | N.A. | ||
| Summary | This gene encodes a member of the MAP kinase family. MAP kinases, also known as extracellular signal-regulated kinases (ERKs), act as an integration point for multiple biochemical signals, and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation and development. The activation of this kinase requires its phosphorylation by upstream kinases. Upon activation, this kinase translocates to the nucleus of the stimulated cells, where it phosphorylates nuclear targets. One study also suggests that this protein acts as a transcriptional repressor independent of its kinase activity. The encoded protein has been identified as a moonlighting protein based on its ability to perform mechanistically distinct functions. Two alternatively spliced transcript variants encoding the same protein, but differing in the UTRs, have been reported for this gene. [provided by RefSeq, Jan 2014] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| MAPK1-201 | ENST00000215832 | 5881 | 360aa | ENSP00000215832 |
| MAPK1-203 | ENST00000491588 | 491 | No protein | - |
| MAPK1-202 | ENST00000398822 | 1487 | 360aa | ENSP00000381803 |
| MAPK1-204 | ENST00000544786 | 951 | 316aa | ENSP00000440842 |
Phenotypes
| ensgID | Trait | pValue | Pubmed ID |
|---|---|---|---|
| 27504 | Cardiovascular Physiological Phenomena | 1.2150000E-005 | - |
| 100307 | Multiple Sclerosis | 5E-9 | 21833088 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
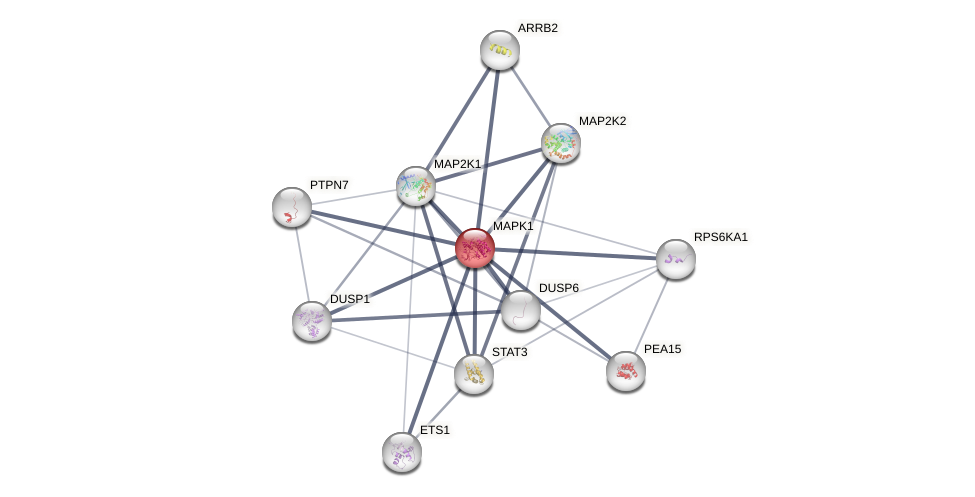
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000100030 | homo_sapiens | ENSP00000215659 | 55.3134 | 37.6022 | homo_sapiens | ENSP00000215832 | 56.3889 | 38.3333 |
| ENSG00000100030 | homo_sapiens | ENSP00000378968 | 29.5343 | 21.201 | homo_sapiens | ENSP00000215832 | 66.9444 | 48.0556 |
| ENSG00000100030 | homo_sapiens | ENSP00000333685 | 57.4176 | 40.3846 | homo_sapiens | ENSP00000215832 | 58.0556 | 40.8333 |
| ENSG00000100030 | homo_sapiens | ENSP00000346484 | 27.0062 | 17.4383 | homo_sapiens | ENSP00000215832 | 48.6111 | 31.3889 |
| ENSG00000100030 | homo_sapiens | ENSP00000501692 | 27.5316 | 18.6709 | homo_sapiens | ENSP00000215832 | 48.3333 | 32.7778 |
| ENSG00000100030 | homo_sapiens | ENSP00000261845 | 29.5423 | 20.111 | homo_sapiens | ENSP00000215832 | 59.1667 | 40.2778 |
| ENSG00000100030 | homo_sapiens | ENSP00000377262 | 21.03 | 13.1617 | homo_sapiens | ENSP00000215832 | 40.8333 | 25.5556 |
| ENSG00000100030 | homo_sapiens | ENSP00000337691 | 37.3162 | 26.6544 | homo_sapiens | ENSP00000215832 | 56.3889 | 40.2778 |
| ENSG00000100030 | homo_sapiens | ENSP00000355304 | 37.9475 | 23.8663 | homo_sapiens | ENSP00000215832 | 44.1667 | 27.7778 |
| ENSG00000100030 | homo_sapiens | ENSP00000384625 | 41.3662 | 27.8937 | homo_sapiens | ENSP00000215832 | 60.5556 | 40.8333 |
| ENSG00000100030 | homo_sapiens | ENSP00000383234 | 35.7751 | 23.8501 | homo_sapiens | ENSP00000215832 | 58.3333 | 38.8889 |
| ENSG00000100030 | homo_sapiens | ENSP00000363304 | 49.6487 | 32.7869 | homo_sapiens | ENSP00000215832 | 58.8889 | 38.8889 |
| ENSG00000100030 | homo_sapiens | ENSP00000493435 | 45.6897 | 30.6034 | homo_sapiens | ENSP00000215832 | 58.8889 | 39.4444 |
| ENSG00000100030 | homo_sapiens | ENSP00000263025 | 88.1266 | 82.3219 | homo_sapiens | ENSP00000215832 | 92.7778 | 86.6667 |
| ENSG00000100030 | homo_sapiens | ENSP00000229794 | 61.1111 | 43.8889 | homo_sapiens | ENSP00000215832 | 61.1111 | 43.8889 |
| ENSG00000100030 | homo_sapiens | ENSP00000362931 | 22.1374 | 14.1985 | homo_sapiens | ENSP00000215832 | 40.2778 | 25.8333 |
| ENSG00000100030 | homo_sapiens | ENSP00000211287 | 55.6164 | 38.9041 | homo_sapiens | ENSP00000215832 | 56.3889 | 39.4444 |
| ENSG00000100030 | homo_sapiens | ENSP00000359119 | 24.8677 | 15.3439 | homo_sapiens | ENSP00000215832 | 39.1667 | 24.1667 |
| ENSG00000100030 | homo_sapiens | ENSP00000394560 | 48.8208 | 32.0755 | homo_sapiens | ENSP00000215832 | 57.5 | 37.7778 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 94.7222 | 94.7222 | nomascus_leucogenys | ENSNLEP00000027273 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 90.2778 | 89.7222 | pan_paniscus | ENSPPAP00000006243 | 95.8702 | 95.2802 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.7222 | 99.7222 | pongo_abelii | ENSPPYP00000023664 | 98.8981 | 98.8981 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | pan_troglodytes | ENSPTRP00000024313 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.0556 | 97.7778 | gorilla_gorilla | ENSGGOP00000040522 | 93.1398 | 92.876 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 86.3889 | 81.6667 | ornithorhynchus_anatinus | ENSOANP00000043143 | 74.0476 | 70 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 91.1111 | 88.8889 | sarcophilus_harrisii | ENSSHAP00000032133 | 94.2529 | 91.954 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 60.5556 | 60.2778 | notamacropus_eugenii | ENSMEUP00000003820 | 60.7242 | 60.4457 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 78.0556 | 77.7778 | choloepus_hoffmanni | ENSCHOP00000008916 | 87.8125 | 87.5 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 82.2222 | 80.5556 | dasypus_novemcinctus | ENSDNOP00000007040 | 90.7975 | 88.9571 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 84.7222 | 81.9444 | dasypus_novemcinctus | ENSDNOP00000035147 | 96.519 | 93.3544 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 73.6111 | 73.6111 | erinaceus_europaeus | ENSEEUP00000002310 | 74.0223 | 74.0223 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.1667 | 98.8889 | echinops_telfairi | ENSETEP00000009702 | 99.4429 | 99.1643 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 65.8333 | 62.2222 | callithrix_jacchus | ENSCJAP00000090447 | 59.6977 | 56.4232 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | cercocebus_atys | ENSCATP00000015951 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | macaca_fascicularis | ENSMFAP00000004754 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | macaca_mulatta | ENSMMUP00000016444 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 95.2778 | macaca_nemestrina | ENSMNEP00000011854 | 96.9014 | 96.6197 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | papio_anubis | ENSPANP00000025866 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 91.6667 | 90.8333 | mandrillus_leucophaeus | ENSMLEP00000023677 | 96.2099 | 95.3353 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 88.8889 | 88.8889 | tursiops_truncatus | ENSTTRP00000015859 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 92.7778 | 92.2222 | vulpes_vulpes | ENSVVUP00000027843 | 85.2041 | 84.6939 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.8889 | 98.6111 | mus_spretus | MGP_SPRETEiJ_P0042419 | 99.4413 | 99.162 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | equus_asinus | ENSEASP00005032621 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 99.7222 | capra_hircus | ENSCHIP00000000204 | 90.9091 | 90.6566 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 88.3333 | 87.7778 | loxodonta_africana | ENSLAFP00000008420 | 96.6565 | 96.0486 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.7222 | 99.4444 | panthera_leo | ENSPLOP00000017103 | 86.506 | 86.2651 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 91.9444 | 90.8333 | ailuropoda_melanoleuca | ENSAMEP00000030976 | 91.1846 | 90.0826 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.7222 | 99.4444 | bos_indicus_hybrid | ENSBIXP00005011080 | 56.6246 | 56.4669 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 90.8333 | 90 | oryctolagus_cuniculus | ENSOCUP00000011783 | 89.8352 | 89.011 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.4444 | 99.4444 | equus_caballus | ENSECAP00000084769 | 60.2694 | 60.2694 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | canis_lupus_familiaris | ENSCAFP00845036530 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 99.4444 | panthera_pardus | ENSPPRP00000017574 | 100 | 99.4444 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 99.7222 | marmota_marmota_marmota | ENSMMMP00000000591 | 100 | 99.7222 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.4444 | 99.4444 | balaenoptera_musculus | ENSBMSP00010020003 | 99.7215 | 99.7215 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 89.1667 | 88.6111 | cavia_porcellus | ENSCPOP00000011740 | 98.7692 | 98.1538 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 99.7222 | felis_catus | ENSFCAP00000025541 | 51.5759 | 51.4327 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 91.9444 | 90.2778 | ursus_americanus | ENSUAMP00000013584 | 90.9341 | 89.2857 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.7778 | 97.5 | monodelphis_domestica | ENSMODP00000022098 | 98.0501 | 97.7716 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 90.5556 | 88.8889 | monodelphis_domestica | ENSMODP00000018334 | 92.3513 | 90.6516 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 91.9444 | 90.5556 | bison_bison_bison | ENSBBBP00000028006 | 88.7399 | 87.3995 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.1667 | 99.1667 | monodon_monoceros | ENSMMNP00015005638 | 99.7207 | 99.7207 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.4444 | 99.4444 | sus_scrofa | ENSSSCP00000073842 | 63.0282 | 63.0282 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | microcebus_murinus | ENSMICP00000017583 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.4444 | 98.8889 | octodon_degus | ENSODEP00000021491 | 99.7215 | 99.1643 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.7222 | 99.4444 | jaculus_jaculus | ENSJJAP00000022529 | 99.7222 | 99.4444 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.7222 | 99.4444 | ovis_aries_rambouillet | ENSOARP00020033914 | 99.446 | 99.169 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 91.9444 | 90.5556 | ursus_maritimus | ENSUMAP00000004216 | 91.1846 | 89.8072 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.4444 | 99.4444 | ochotona_princeps | ENSOPRP00000013218 | 99.7215 | 99.7215 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.1667 | 99.1667 | delphinapterus_leucas | ENSDLEP00000021680 | 93.9474 | 93.9474 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.4444 | 99.4444 | physeter_catodon | ENSPCTP00005019452 | 99.7215 | 99.7215 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.8889 | 98.6111 | mus_caroli | MGP_CAROLIEiJ_P0040564 | 99.4413 | 99.162 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.8889 | 98.6111 | mus_musculus | ENSMUSP00000065983 | 99.4413 | 99.162 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 99.4444 | dipodomys_ordii | ENSDORP00000016925 | 100 | 99.4444 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 89.7222 | 89.1667 | mustela_putorius_furo | ENSMPUP00000003112 | 96.997 | 96.3964 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.8889 | 98.8889 | aotus_nancymaae | ENSANAP00000014877 | 94.6809 | 94.6809 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 81.9444 | 81.3889 | saimiri_boliviensis_boliviensis | ENSSBOP00000023855 | 96.7213 | 96.0656 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | chlorocebus_sabaeus | ENSCSAP00000009828 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 61.3889 | 61.1111 | tupaia_belangeri | ENSTBEP00000007776 | 85.6589 | 85.2713 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.8889 | 98.6111 | mesocricetus_auratus | ENSMAUP00000018564 | 99.4413 | 99.162 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | phocoena_sinus | ENSPSNP00000018097 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.4444 | 99.4444 | camelus_dromedarius | ENSCDRP00005032276 | 99.7215 | 99.7215 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.7222 | 99.1667 | carlito_syrichta | ENSTSYP00000032180 | 98.8981 | 98.3471 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 88.8889 | 88.6111 | panthera_tigris_altaica | ENSPTIP00000004736 | 88.8889 | 88.6111 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.5 | 96.9444 | rattus_norvegicus | ENSRNOP00000086878 | 94.3548 | 93.8172 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | prolemur_simus | ENSPSMP00000020060 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.6111 | 98.3333 | microtus_ochrogaster | ENSMOCP00000012005 | 99.4398 | 99.1597 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 62.5 | 62.2222 | sorex_araneus | ENSSARP00000013061 | 79.7872 | 79.4326 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.6111 | 98.3333 | peromyscus_maniculatus_bairdii | ENSPEMP00000012990 | 99.4398 | 99.1597 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 90.5556 | 89.7222 | bos_mutus | ENSBMUP00000003546 | 98.1928 | 97.2892 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.8889 | 98.6111 | mus_spicilegus | ENSMSIP00000020806 | 99.4413 | 99.162 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.8889 | 98.6111 | cricetulus_griseus_chok1gshd | ENSCGRP00001022860 | 99.4413 | 99.162 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 88.6111 | 88.6111 | pteropus_vampyrus | ENSPVAP00000012866 | 88.8579 | 88.8579 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 88.8889 | 88.8889 | myotis_lucifugus | ENSMLUP00000005441 | 99.6885 | 99.6885 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 93.3333 | 92.5 | myotis_lucifugus | ENSMLUP00000022195 | 93.3333 | 92.5 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.1111 | 95.8333 | vombatus_ursinus | ENSVURP00010028502 | 99.1404 | 98.8539 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | rhinolophus_ferrumequinum | ENSRFEP00010026783 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | neovison_vison | ENSNVIP00000027883 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 99.7222 | bos_taurus | ENSBTAP00000013623 | 100 | 99.7222 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 99.7222 | heterocephalus_glaber_female | ENSHGLP00000029496 | 100 | 99.7222 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95 | 95 | rhinopithecus_bieti | ENSRBIP00000026426 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.6667 | 96.6667 | canis_lupus_dingo | ENSCAFP00020019010 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 99.7222 | sciurus_vulgaris | ENSSVLP00005008763 | 100 | 99.7222 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 84.1667 | 80.8333 | phascolarctos_cinereus | ENSPCIP00000035808 | 85.8357 | 82.4363 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 81.3889 | 81.3889 | procavia_capensis | ENSPCAP00000009794 | 81.6156 | 81.6156 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.6111 | 98.3333 | nannospalax_galili | ENSNGAP00000016309 | 98.6111 | 98.3333 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.7222 | 99.4444 | bos_grunniens | ENSBGRP00000039366 | 56.5354 | 56.378 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 99.7222 | chinchilla_lanigera | ENSCLAP00000024734 | 100 | 99.7222 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 83.0556 | 81.6667 | cervus_hanglu_yarkandensis | ENSCHYP00000009688 | 73.8272 | 72.5926 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.4444 | 99.4444 | catagonus_wagneri | ENSCWAP00000008234 | 99.7215 | 99.7215 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 90.2778 | 89.1667 | ictidomys_tridecemlineatus | ENSSTOP00000013254 | 98.1873 | 96.9789 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 88.6111 | 88.6111 | otolemur_garnettii | ENSOGAP00000008157 | 98.7616 | 98.7616 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.5 | 97.2222 | cebus_imitator | ENSCCAP00000014631 | 95.9016 | 95.6284 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 99.7222 | urocitellus_parryii | ENSUPAP00010017096 | 100 | 99.7222 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 99.7222 | 99.4444 | moschus_moschiferus | ENSMMSP00000021811 | 99.1713 | 98.895 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 89.7222 | 88.8889 | rhinopithecus_roxellana | ENSRROP00000024767 | 97.8788 | 96.9697 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.8889 | 98.6111 | mus_pahari | MGP_PahariEiJ_P0027958 | 99.4413 | 99.162 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 100 | 100 | propithecus_coquereli | ENSPCOP00000029858 | 100 | 100 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 85 | 77.2222 | caenorhabditis_elegans | F43C1.2b.1 | 68.9189 | 62.6126 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 86.6667 | 78.6111 | drosophila_melanogaster | FBpp0311493 | 82.9787 | 75.266 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 87.5 | 80.5556 | ciona_intestinalis | ENSCINP00000014552 | 90 | 82.8571 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 94.4444 | 89.1667 | petromyzon_marinus | ENSPMAP00000010177 | 91.3979 | 86.2903 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 68.3333 | 64.7222 | eptatretus_burgeri | ENSEBUP00000017381 | 92.8302 | 87.9245 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 94.7222 | 91.6667 | callorhinchus_milii | ENSCMIP00000032576 | 87.6607 | 84.8329 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.6667 | 93.3333 | latimeria_chalumnae | ENSLACP00000015867 | 95.3425 | 92.0548 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.3889 | 93.3333 | lepisosteus_oculatus | ENSLOCP00000002360 | 94.0379 | 91.0569 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95 | 90.5556 | clupea_harengus | ENSCHAP00000058109 | 87.9177 | 83.8046 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.7778 | danio_rerio | ENSDARP00000038550 | 93.2249 | 90.5149 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 94.7222 | 92.2222 | carassius_auratus | ENSCARP00000037792 | 87.4359 | 85.1282 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95 | 91.6667 | carassius_auratus | ENSCARP00000124448 | 87.9177 | 84.8329 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 88.8889 | 84.7222 | astyanax_mexicanus | ENSAMXP00000001246 | 78.0488 | 74.3902 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95 | 92.7778 | ictalurus_punctatus | ENSIPUP00000030101 | 91.2 | 89.0667 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 93.8889 | 90.5556 | esox_lucius | ENSELUP00000069352 | 88.7139 | 85.5643 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.2778 | 92.2222 | electrophorus_electricus | ENSEEEP00000018510 | 88.8601 | 86.0104 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 67.7778 | 57.5 | oncorhynchus_kisutch | ENSOKIP00005028637 | 35.2601 | 29.9133 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 93.6111 | 90.8333 | oncorhynchus_kisutch | ENSOKIP00005009666 | 91.3279 | 88.6179 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 93.6111 | 90.8333 | oncorhynchus_mykiss | ENSOMYP00000129285 | 88.4514 | 85.8268 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 91.1111 | 88.6111 | oncorhynchus_mykiss | ENSOMYP00000132444 | 89.6175 | 87.1585 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 93.8889 | 90.5556 | salmo_salar | ENSSSAP00000130067 | 91.5989 | 88.3469 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 91.9444 | 88.8889 | salmo_salar | ENSSSAP00000162170 | 76.7981 | 74.2459 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 71.9444 | 69.4444 | salmo_salar | ENSSSAP00000173949 | 93.8406 | 90.5797 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 86.9444 | 82.2222 | salmo_trutta | ENSSTUP00000017302 | 55.2028 | 52.2046 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 93.6111 | 90.8333 | salmo_trutta | ENSSTUP00000030531 | 89.6277 | 86.9681 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.2778 | 91.3889 | gadus_morhua | ENSGMOP00000014982 | 88.4021 | 84.7938 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95 | 91.9444 | fundulus_heteroclitus | ENSFHEP00000011839 | 92.6829 | 89.7019 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.1111 | 92.5 | poecilia_reticulata | ENSPREP00000019735 | 95.0549 | 91.4835 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.1111 | 92.5 | xiphophorus_maculatus | ENSXMAP00000006468 | 95.0549 | 91.4835 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.5 | oryzias_latipes | ENSORLP00000039541 | 93.2249 | 90.2439 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 88.0556 | 83.8889 | cyclopterus_lumpus | ENSCLMP00005032781 | 76.57 | 72.9469 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.8333 | 92.2222 | oreochromis_niloticus | ENSONIP00000077238 | 93.4959 | 89.9729 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.8333 | 92.2222 | haplochromis_burtoni | ENSHBUP00000024696 | 93.4959 | 89.9729 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.8333 | 92.2222 | astatotilapia_calliptera | ENSACLP00000005434 | 93.4959 | 89.9729 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.2778 | 91.6667 | sparus_aurata | ENSSAUP00010010405 | 90.5013 | 87.0712 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 86.6667 | 82.7778 | lates_calcarifer | ENSLCAP00010051733 | 82.1053 | 78.4211 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.9444 | 95.5556 | xenopus_tropicalis | ENSXETP00000079571 | 96.6759 | 95.2909 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.7778 | 96.9444 | chrysemys_picta_bellii | ENSCPBP00000014183 | 95.6522 | 94.837 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.0556 | 96.6667 | sphenodon_punctatus | ENSSPUP00000020365 | 95.9239 | 94.5652 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 75.5556 | 73.0556 | crocodylus_porosus | ENSCPRP00005000848 | 91.2752 | 88.255 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.5 | 94.7222 | laticauda_laticaudata | ENSLLTP00000008424 | 95.3804 | 92.663 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.9444 | 94.4444 | notechis_scutatus | ENSNSUP00000012703 | 95.6164 | 93.1507 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.2222 | 94.4444 | pseudonaja_textilis | ENSPTXP00000011774 | 95.1087 | 92.3913 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.7778 | 97.2222 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000005723 | 95.6522 | 95.1087 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 93.8889 | 90.8333 | gallus_gallus | ENSGALP00010036277 | 80.285 | 77.6722 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 91.9444 | 89.7222 | meleagris_gallopavo | ENSMGAP00000000469 | 88.2667 | 86.1333 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.3889 | 96.1111 | serinus_canaria | ENSSCAP00000005913 | 99.7126 | 99.4253 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.3333 | 97.5 | parus_major | ENSPMJP00000019015 | 76.7896 | 76.1388 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.2222 | 95 | anolis_carolinensis | ENSACAP00000001346 | 95.1087 | 92.9348 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 78.8889 | 75 | neolamprologus_brichardi | ENSNBRP00000023207 | 83.0409 | 78.9474 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.2222 | 94.7222 | naja_naja | ENSNNAP00000017484 | 95.1087 | 92.663 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.7778 | 95.5556 | podarcis_muralis | ENSPMRP00000029862 | 95.6522 | 93.4783 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 89.7222 | 89.4444 | geospiza_fortis | ENSGFOP00000000286 | 95.5621 | 95.2663 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.0556 | 96.9444 | taeniopygia_guttata | ENSTGUP00000031436 | 95.664 | 94.5799 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 90.8333 | erpetoichthys_calabaricus | ENSECRP00000029456 | 91.9786 | 87.4332 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 93.8889 | 90.2778 | kryptolebias_marmoratus | ENSKMAP00000005892 | 91.5989 | 88.0759 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.1111 | 95 | salvator_merianae | ENSSMRP00000012334 | 94.0217 | 92.9348 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 86.9444 | 83.8889 | oryzias_javanicus | ENSOJAP00000002096 | 91.7889 | 88.563 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 84.1667 | 81.1111 | amphilophus_citrinellus | ENSACIP00000007539 | 93.2308 | 89.8462 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 91.9444 | dicentrarchus_labrax | ENSDLAP00005058881 | 88.6598 | 85.3093 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | amphiprion_percula | ENSAPEP00000026166 | 93.2249 | 89.9729 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 43.6111 | 42.2222 | oryzias_sinensis | ENSOSIP00000006215 | 96.9136 | 93.8272 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 88.6111 | 87.2222 | pelodiscus_sinensis | ENSPSIP00000003362 | 99.0683 | 97.5155 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.7778 | 96.9444 | gopherus_evgoodei | ENSGEVP00005014698 | 84.0095 | 83.2936 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.7778 | 96.6667 | chelonoidis_abingdonii | ENSCABP00000028265 | 94.37 | 93.2976 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | seriola_dumerili | ENSSDUP00000013074 | 93.2249 | 89.9729 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95 | 91.1111 | cottoperca_gobio | ENSCGOP00000056359 | 87.6923 | 84.1026 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 84.7222 | 82.2222 | struthio_camelus_australis | ENSSCUP00000005553 | 82.2102 | 79.7844 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 89.4444 | 88.6111 | anser_brachyrhynchus | ENSABRP00000016589 | 96.4072 | 95.509 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 94.7222 | 91.1111 | gasterosteus_aculeatus | ENSGACP00000019049 | 92.4119 | 88.8889 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 92.7778 | 88.6111 | cynoglossus_semilaevis | ENSCSEP00000029876 | 93.2961 | 89.1061 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 34.4444 | 30 | cynoglossus_semilaevis | ENSCSEP00000011950 | 72.093 | 62.7907 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.3889 | 92.7778 | poecilia_formosa | ENSPFOP00000006223 | 95.3297 | 91.7582 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 40.5556 | 37.7778 | myripristis_murdjan | ENSMMDP00005009187 | 96.6887 | 90.0662 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.2778 | 91.9444 | sander_lucioperca | ENSSLUP00000036965 | 87.2774 | 84.2239 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.0556 | 96.9444 | strigops_habroptila | ENSSHBP00005010516 | 95.9239 | 94.837 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | amphiprion_ocellaris | ENSAOCP00000001430 | 93.2249 | 89.9729 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.2222 | 96.3889 | terrapene_carolina_triunguis | ENSTMTP00000009038 | 95.6284 | 94.8087 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 20.2778 | 19.4444 | hucho_hucho | ENSHHUP00000006905 | 96.0526 | 92.1053 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 62.7778 | 56.6667 | hucho_hucho | ENSHHUP00000028574 | 76.8708 | 69.3878 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.5 | betta_splendens | ENSBSLP00000041845 | 89.3506 | 86.4935 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.8333 | 92.7778 | pygocentrus_nattereri | ENSPNAP00000032622 | 92.9919 | 90.027 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | anabas_testudineus | ENSATEP00000058109 | 87.9795 | 84.9105 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 86.9444 | 80.8333 | ciona_savignyi | ENSCSAVP00000014424 | 89.4286 | 83.1429 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 60 | 40.2778 | saccharomyces_cerevisiae | YKL161C | 49.8845 | 33.4873 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 65 | 46.6667 | saccharomyces_cerevisiae | YHR030C | 48.3471 | 34.7107 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | seriola_lalandi_dorsalis | ENSSLDP00000033321 | 93.2249 | 89.9729 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 94.7222 | 90.8333 | labrus_bergylta | ENSLBEP00000023541 | 91.9137 | 88.1402 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 85.8333 | 81.6667 | pundamilia_nyererei | ENSPNYP00000007060 | 92.515 | 88.0239 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | mastacembelus_armatus | ENSMAMP00000023547 | 87.3096 | 84.264 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | stegastes_partitus | ENSSPAP00000031074 | 93.2249 | 89.9729 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 74.4444 | 70.2778 | oncorhynchus_tshawytscha | ENSOTSP00005053383 | 81.7073 | 77.1341 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 35.8333 | 34.1667 | acanthochromis_polyacanthus | ENSAPOP00000027566 | 86 | 82 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | acanthochromis_polyacanthus | ENSAPOP00000010041 | 93.2249 | 89.9729 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 80.2778 | 76.9444 | acanthochromis_polyacanthus | ENSAPOP00000028293 | 92.3323 | 88.4984 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 98.0556 | 97.5 | aquila_chrysaetos_chrysaetos | ENSACCP00020005442 | 95.9239 | 95.3804 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.8333 | 92.5 | nothobranchius_furzeri | ENSNFUP00015011993 | 88.9175 | 85.8247 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 90.2778 | 86.1111 | cyprinodon_variegatus | ENSCVAP00000015363 | 92.5926 | 88.3191 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95 | 90.5556 | denticeps_clupeoides | ENSDCDP00000049874 | 88.601 | 84.456 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | larimichthys_crocea | ENSLCRP00005016597 | 87.5318 | 84.4784 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | takifugu_rubripes | ENSTRUP00000051554 | 88.2051 | 85.1282 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 91.9444 | 89.7222 | ficedula_albicollis | ENSFALP00000008282 | 83.7975 | 81.7721 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 91.9444 | hippocampus_comes | ENSHCOP00000021531 | 93.4783 | 89.9457 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.2778 | 92.2222 | cyprinus_carpio_carpio | ENSCCRP00000000238 | 83.8631 | 81.1736 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95 | 92.7778 | cyprinus_carpio_carpio | ENSCCRP00000046771 | 88.1443 | 86.0825 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 97.7778 | 97.2222 | coturnix_japonica | ENSCJPP00005024064 | 95.6522 | 95.1087 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 96.3889 | 93.0556 | poecilia_latipinna | ENSPLAP00000016708 | 95.3297 | 92.033 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 94.7222 | 91.9444 | sinocyclocheilus_grahami | ENSSGRP00000074851 | 87.8866 | 85.3093 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95 | 92.7778 | sinocyclocheilus_grahami | ENSSGRP00000087334 | 88.8312 | 86.7533 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.8333 | 92.2222 | maylandia_zebra | ENSMZEP00005027661 | 93.4959 | 89.9729 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.8333 | 91.6667 | tetraodon_nigroviridis | ENSTNIP00000018415 | 94.0054 | 89.9183 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 90.8333 | paramormyrops_kingsleyae | ENSPKIP00000033096 | 93.2249 | 88.6179 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | scleropages_formosus | ENSSFOP00015047026 | 85.1485 | 82.1782 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 90.8333 | 89.1667 | leptobrachium_leishanense | ENSLLEP00000014533 | 81.1414 | 79.6526 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.5556 | 92.2222 | scophthalmus_maximus | ENSSMAP00000037872 | 79.4457 | 76.6744 |
| ENSG00000100030 | homo_sapiens | ENSP00000215832 | 95.8333 | 92.7778 | oryzias_melastigma | ENSOMEP00000008061 | 93.4959 | 90.5149 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0001784 | enables phosphotyrosine residue binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0003677 | enables DNA binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0004674 | enables protein serine/threonine kinase activity | 2 | IBA,IDA | Homo_sapiens(9606) | Function |
| GO:0004707 | enables MAP kinase activity | 1 | TAS | Homo_sapiens(9606) | Function |
| GO:0004708 | enables MAP kinase kinase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005524 | enables ATP binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005576 | located_in extracellular region | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005634 | located_in nucleus | 4 | HDA,IBA,IDA,TAS | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 4 | HDA,IBA,IDA,ISS | Homo_sapiens(9606) | Component |
| GO:0005739 | located_in mitochondrion | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005769 | located_in early endosome | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005770 | located_in late endosome | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005788 | located_in endoplasmic reticulum lumen | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005794 | located_in Golgi apparatus | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005815 | located_in microtubule organizing center | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0005829 | located_in cytosol | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005856 | located_in cytoskeleton | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005886 | located_in plasma membrane | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0005901 | located_in caveola | 2 | ISS,TAS | Homo_sapiens(9606) | Component |
| GO:0005925 | located_in focal adhesion | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0006357 | involved_in regulation of transcription by RNA polymerase II | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006468 | involved_in protein phosphorylation | 2 | IDA,TAS | Homo_sapiens(9606) | Process |
| GO:0006915 | involved_in apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006935 | involved_in chemotaxis | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0006974 | involved_in DNA damage response | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007049 | involved_in cell cycle | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007165 | involved_in signal transduction | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007166 | involved_in cell surface receptor signaling pathway | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0007268 | involved_in chemical synaptic transmission | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0007611 | involved_in learning or memory | 1 | NAS | Homo_sapiens(9606) | Process |
| GO:0008286 | involved_in insulin receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0008353 | enables RNA polymerase II CTD heptapeptide repeat kinase activity | 1 | ISS | Homo_sapiens(9606) | Function |
| GO:0010759 | acts_upstream_of positive regulation of macrophage chemotaxis | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:0010800 | involved_in positive regulation of peptidyl-threonine phosphorylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0018105 | involved_in peptidyl-serine phosphorylation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0018107 | involved_in peptidyl-threonine phosphorylation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0019858 | involved_in cytosine metabolic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0019902 | enables phosphatase binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0030278 | involved_in regulation of ossification | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030521 | involved_in androgen receptor signaling pathway | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0030641 | involved_in regulation of cellular pH | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0030878 | involved_in thyroid gland development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0031143 | located_in pseudopodium | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0031647 | involved_in regulation of protein stability | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0031663 | involved_in lipopolysaccharide-mediated signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0032212 | involved_in positive regulation of telomere maintenance via telomerase | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032872 | involved_in regulation of stress-activated MAPK cascade | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0033598 | involved_in mammary gland epithelial cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0034198 | involved_in cellular response to amino acid starvation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0034614 | involved_in cellular response to reactive oxygen species | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0035094 | involved_in response to nicotine | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0035556 | involved_in intracellular signal transduction | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0035578 | located_in azurophil granule lumen | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0038127 | involved_in ERBB signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0042473 | involved_in outer ear morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0042802 | enables identical protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0043330 | involved_in response to exogenous dsRNA | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043401 | involved_in steroid hormone mediated signaling pathway | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0045202 | located_in synapse | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0045596 | involved_in negative regulation of cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048009 | involved_in insulin-like growth factor receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0048538 | involved_in thymus development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0050847 | involved_in progesterone receptor signaling pathway | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0050852 | involved_in T cell receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0050853 | involved_in B cell receptor signaling pathway | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0051403 | involved_in stress-activated MAPK cascade | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0051493 | involved_in regulation of cytoskeleton organization | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0051973 | involved_in positive regulation of telomerase activity | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0060020 | involved_in Bergmann glial cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060291 | involved_in long-term synaptic potentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060324 | involved_in face development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060425 | involved_in lung morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060440 | involved_in trachea formation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060716 | involved_in labyrinthine layer blood vessel development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0061308 | involved_in cardiac neural crest cell development involved in heart development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0070371 | involved_in ERK1 and ERK2 cascade | 2 | IC,IDA | Homo_sapiens(9606) | Process |
| GO:0070849 | involved_in response to epidermal growth factor | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071276 | involved_in cellular response to cadmium ion | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0071356 | involved_in cellular response to tumor necrosis factor | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0072584 | involved_in caveolin-mediated endocytosis | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0072686 | located_in mitotic spindle | 1 | ISS | Homo_sapiens(9606) | Component |
| GO:0090170 | involved_in regulation of Golgi inheritance | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0106310 | enables protein serine kinase activity | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0120041 | acts_upstream_of positive regulation of macrophage proliferation | 1 | IGI | Homo_sapiens(9606) | Process |
| GO:1904355 | involved_in positive regulation of telomere capping | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1904813 | located_in ficolin-1-rich granule lumen | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:2000641 | involved_in regulation of early endosome to late endosome transport | 1 | TAS | Homo_sapiens(9606) | Process |