Welcome to
RBP World!
Description
| Ensembl ID | ENSG00000019549 | Gene ID | 6591 | Accession | 11094 |
| Symbol | SNAI2 | Alias | SLUG;WS2D;SLUGH;SLUGH1;SNAIL2 | Full Name | snail family transcriptional repressor 2 |
| Status | Confidence | Length | 4143 bases | Strand | Minus strand |
| Position | 8 : 48917598 - 48921740 | RNA binding domain | zf-met | ||
| Summary | This gene encodes a member of the Snail family of C2H2-type zinc finger transcription factors. The encoded protein acts as a transcriptional repressor that binds to E-box motifs and is also likely to repress E-cadherin transcription in breast carcinoma. This protein is involved in epithelial-mesenchymal transitions and has antiapoptotic activity. Mutations in this gene may be associated with sporatic cases of neural tube defects. [provided by RefSeq, Jul 2008] | ||||
RNA binding domains (RBDs)
| Protein | Domain | Pfam ID | E-value | Domain number |
|---|---|---|---|---|
| ENSP00000020945 | zf-met | PF12874 | 3.4e-07 | 1 |
RNA binding proteomes (RBPomes)
| Pubmed ID | Full Name | Cell | Author | Time | Doi |
|---|
Literatures on RNA binding capacity
| Pubmed ID | Title | Author | Time | Journal |
|---|---|---|---|---|
| 30602079 | Roles of Oncogenic Long Non-coding RNAs in Cancer Development. | Hyunhee Do | 2018-12-01 | Genomics & informatics |
| 28954219 | The Cohesin Complex Is Necessary for Epidermal Progenitor Cell Function through Maintenance of Self-Renewal Genes. | Maria Noutsou | 2017-09-26 | Cell reports |
| 33420019 | Interaction between SNAI2 and MYOD enhances oncogenesis and suppresses differentiation in Fusion Negative Rhabdomyosarcoma. | Silvia Pomella | 2021-01-08 | Nature communications |
Transcripts
| Name | Transcript ID | bp | Protein | Translation ID |
|---|---|---|---|---|
| SNAI2-201 | ENST00000020945 | 2180 | 268aa | ENSP00000020945 |
| SNAI2-203 | ENST00000649776 | 2828 | No protein | - |
| SNAI2-202 | ENST00000396822 | 626 | 89aa | ENSP00000380034 |
GWAS
| ensgID | SNP | Chromosome | Position | Trait | PubmedID | Or or BEAT | EFO ID |
|---|
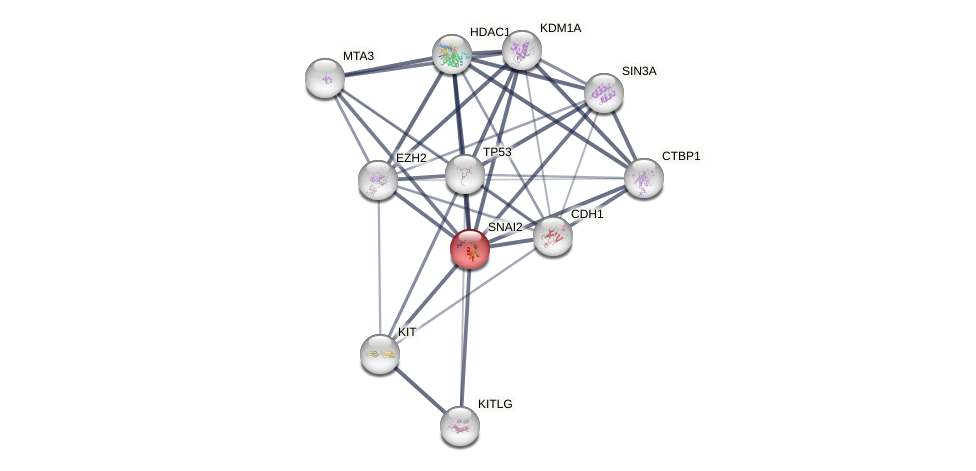
Protein-Protein Interaction (PPI)
Paralogs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000019549 | homo_sapiens | ENSP00000327968 | 56.8493 | 46.5753 | homo_sapiens | ENSP00000020945 | 61.9403 | 50.7463 |
| ENSG00000019549 | homo_sapiens | ENSP00000455711 | 38.7931 | 29.5977 | homo_sapiens | ENSP00000020945 | 50.3731 | 38.4328 |
| ENSG00000019549 | homo_sapiens | ENSP00000246104 | 41.3681 | 32.899 | homo_sapiens | ENSP00000020945 | 47.3881 | 37.6866 |
| ENSG00000019549 | homo_sapiens | ENSP00000338157 | 14.8588 | 9.21248 | homo_sapiens | ENSP00000020945 | 37.3134 | 23.1343 |
| ENSG00000019549 | homo_sapiens | ENSP00000315870 | 29.3878 | 20.4082 | homo_sapiens | ENSP00000020945 | 26.8657 | 18.6567 |
| ENSG00000019549 | homo_sapiens | ENSP00000276594 | 16.1121 | 10.683 | homo_sapiens | ENSP00000020945 | 34.3284 | 22.7612 |
| ENSG00000019549 | homo_sapiens | ENSP00000376035 | 13.963 | 8.62423 | homo_sapiens | ENSP00000020945 | 25.3731 | 15.6716 |
| ENSG00000019549 | homo_sapiens | ENSP00000293771 | 15.122 | 8.45528 | homo_sapiens | ENSP00000020945 | 34.7015 | 19.403 |
| ENSG00000019549 | homo_sapiens | ENSP00000358092 | 12.6061 | 7.75758 | homo_sapiens | ENSP00000020945 | 38.806 | 23.8806 |
| ENSG00000019549 | homo_sapiens | ENSP00000359740 | 8.48256 | 4.24128 | homo_sapiens | ENSP00000020945 | 33.5821 | 16.791 |
| ENSG00000019549 | homo_sapiens | ENSP00000379847 | 12.3824 | 8.15047 | homo_sapiens | ENSP00000020945 | 29.4776 | 19.403 |
| ENSG00000019549 | homo_sapiens | ENSP00000351539 | 14.5009 | 10.1695 | homo_sapiens | ENSP00000020945 | 28.7313 | 20.1493 |
| ENSG00000019549 | homo_sapiens | ENSP00000384725 | 14.6218 | 9.91597 | homo_sapiens | ENSP00000020945 | 32.4627 | 22.0149 |
| ENSG00000019549 | homo_sapiens | ENSP00000446335 | 21.6667 | 14.0476 | homo_sapiens | ENSP00000020945 | 33.9552 | 22.0149 |
| ENSG00000019549 | homo_sapiens | ENSP00000305483 | 16.5703 | 9.44123 | homo_sapiens | ENSP00000020945 | 32.0896 | 18.2836 |
| ENSG00000019549 | homo_sapiens | ENSP00000362127 | 12.085 | 7.17131 | homo_sapiens | ENSP00000020945 | 33.9552 | 20.1493 |
| ENSG00000019549 | homo_sapiens | ENSP00000477858 | 12.465 | 8.54342 | homo_sapiens | ENSP00000020945 | 33.209 | 22.7612 |
| ENSG00000019549 | homo_sapiens | ENSP00000302770 | 10.3158 | 6.94737 | homo_sapiens | ENSP00000020945 | 18.2836 | 12.3134 |
| ENSG00000019549 | homo_sapiens | ENSP00000356322 | 8.37989 | 4.80447 | homo_sapiens | ENSP00000020945 | 27.9851 | 16.0448 |
| ENSG00000019549 | homo_sapiens | ENSP00000353863 | 8.43829 | 5.16373 | homo_sapiens | ENSP00000020945 | 25 | 15.2985 |
| ENSG00000019549 | homo_sapiens | ENSP00000363064 | 12.9825 | 8.42105 | homo_sapiens | ENSP00000020945 | 27.6119 | 17.9104 |
| ENSG00000019549 | homo_sapiens | ENSP00000501561 | 12.1457 | 7.01754 | homo_sapiens | ENSP00000020945 | 33.5821 | 19.403 |
| ENSG00000019549 | homo_sapiens | ENSP00000467877 | 15.1858 | 9.2084 | homo_sapiens | ENSP00000020945 | 35.0746 | 21.2687 |
| ENSG00000019549 | homo_sapiens | ENSP00000266269 | 12.8093 | 8.87918 | homo_sapiens | ENSP00000020945 | 32.8358 | 22.7612 |
| ENSG00000019549 | homo_sapiens | ENSP00000507462 | 14.9671 | 9.21053 | homo_sapiens | ENSP00000020945 | 33.9552 | 20.8955 |
| ENSG00000019549 | homo_sapiens | ENSP00000462337 | 15.1631 | 10.7486 | homo_sapiens | ENSP00000020945 | 29.4776 | 20.8955 |
| ENSG00000019549 | homo_sapiens | ENSP00000451763 | 15.0628 | 9.41423 | homo_sapiens | ENSP00000020945 | 26.8657 | 16.791 |
| ENSG00000019549 | homo_sapiens | ENSP00000313362 | 16.5618 | 11.74 | homo_sapiens | ENSP00000020945 | 29.4776 | 20.8955 |
| ENSG00000019549 | homo_sapiens | ENSP00000339030 | 13.1579 | 8.59649 | homo_sapiens | ENSP00000020945 | 27.9851 | 18.2836 |
| ENSG00000019549 | homo_sapiens | ENSP00000409107 | 17.2986 | 13.2701 | homo_sapiens | ENSP00000020945 | 27.2388 | 20.8955 |
| ENSG00000019549 | homo_sapiens | ENSP00000506911 | 10.5189 | 6.31136 | homo_sapiens | ENSP00000020945 | 27.9851 | 16.791 |
| ENSG00000019549 | homo_sapiens | ENSP00000354118 | 8.82353 | 4.84429 | homo_sapiens | ENSP00000020945 | 38.0597 | 20.8955 |
| ENSG00000019549 | homo_sapiens | ENSP00000385319 | 14.7967 | 10.7317 | homo_sapiens | ENSP00000020945 | 33.9552 | 24.6269 |
| ENSG00000019549 | homo_sapiens | ENSP00000244050 | 67.4242 | 55.303 | homo_sapiens | ENSP00000020945 | 66.4179 | 54.4776 |
| ENSG00000019549 | homo_sapiens | ENSP00000469089 | 15.2642 | 9.19765 | homo_sapiens | ENSP00000020945 | 29.1045 | 17.5373 |
| ENSG00000019549 | homo_sapiens | ENSP00000415836 | 14.9837 | 8.30619 | homo_sapiens | ENSP00000020945 | 34.3284 | 19.0299 |
Orthologs
| Ensembl ID | Source | Target | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Protein ID | Perc_pos | Perc_id | Species | Protein ID | Perc_pos | Perc_id | |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 99.6269 | 99.2537 | nomascus_leucogenys | ENSNLEP00000016248 | 99.6269 | 99.2537 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | pan_paniscus | ENSPPAP00000003717 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | pongo_abelii | ENSPPYP00000020834 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | pan_troglodytes | ENSPTRP00000034651 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | gorilla_gorilla | ENSGGOP00000000945 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 96.2687 | 94.0299 | ornithorhynchus_anatinus | ENSOANP00000011569 | 96.2687 | 94.0299 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 97.3881 | 95.1493 | sarcophilus_harrisii | ENSSHAP00000039980 | 97.3881 | 95.1493 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 94.7761 | 92.1642 | notamacropus_eugenii | ENSMEUP00000008671 | 97.318 | 94.636 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 57.4627 | 57.0896 | erinaceus_europaeus | ENSEEUP00000013272 | 96.8553 | 96.2264 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 43.2836 | 39.5522 | echinops_telfairi | ENSETEP00000013276 | 82.2695 | 75.1773 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 94.403 | 92.1642 | callithrix_jacchus | ENSCJAP00000089507 | 81.0897 | 79.1667 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | cercocebus_atys | ENSCATP00000009446 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | macaca_fascicularis | ENSMFAP00000053300 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | macaca_mulatta | ENSMMUP00000021280 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | macaca_nemestrina | ENSMNEP00000020837 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | papio_anubis | ENSPANP00000005532 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | mandrillus_leucophaeus | ENSMLEP00000020399 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 98.1343 | vulpes_vulpes | ENSVVUP00000024020 | 98.5075 | 98.1343 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.1343 | 95.8955 | mus_spretus | MGP_SPRETEiJ_P0042350 | 97.7695 | 95.539 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 95.8955 | 93.2836 | capra_hircus | ENSCHIP00000029179 | 95.8955 | 93.2836 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.1343 | 96.2687 | loxodonta_africana | ENSLAFP00000007793 | 98.1343 | 96.2687 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 97.3881 | panthera_leo | ENSPLOP00000002792 | 98.5075 | 97.3881 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.8806 | 98.5075 | ailuropoda_melanoleuca | ENSAMEP00000010096 | 98.8806 | 98.5075 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 90.2985 | 86.9403 | bos_indicus_hybrid | ENSBIXP00005041936 | 87.0504 | 83.813 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.1343 | 97.7612 | canis_lupus_familiaris | ENSCAFP00845039473 | 98.1343 | 97.7612 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 97.7612 | panthera_pardus | ENSPPRP00000021887 | 98.5075 | 97.7612 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 97.3881 | marmota_marmota_marmota | ENSMMMP00000025345 | 98.5075 | 97.3881 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 97.0149 | 95.5224 | balaenoptera_musculus | ENSBMSP00010028931 | 97.0149 | 95.5224 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 85.0746 | 84.7015 | ursus_americanus | ENSUAMP00000018871 | 99.1304 | 98.6957 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 97.0149 | 94.7761 | monodelphis_domestica | ENSMODP00000012734 | 97.0149 | 94.7761 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 95.8955 | 93.6567 | bison_bison_bison | ENSBBBP00000010656 | 95.8955 | 93.6567 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 97.3881 | 96.2687 | monodon_monoceros | ENSMMNP00015023692 | 97.3881 | 96.2687 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 97.0149 | 95.1493 | sus_scrofa | ENSSSCP00000037603 | 97.0149 | 95.1493 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 95.8955 | 93.2836 | ovis_aries_rambouillet | ENSOARP00020031857 | 95.8955 | 93.2836 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 97.0149 | 96.2687 | ursus_maritimus | ENSUMAP00000008544 | 98.8593 | 98.0989 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 69.7761 | 67.9104 | ochotona_princeps | ENSOPRP00000011541 | 90.7767 | 88.3495 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 91.4179 | 89.1791 | delphinapterus_leucas | ENSDLEP00000021284 | 90.0735 | 87.8676 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 96.6418 | 95.1493 | physeter_catodon | ENSPCTP00005015891 | 96.6418 | 95.1493 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 96.2687 | mus_caroli | MGP_CAROLIEiJ_P0040496 | 98.1413 | 95.9108 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.1343 | 95.5224 | mus_musculus | ENSMUSP00000023356 | 97.7695 | 95.1673 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 96.2687 | 94.403 | dipodomys_ordii | ENSDORP00000018692 | 98.8506 | 96.9349 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.8806 | 98.5075 | mustela_putorius_furo | ENSMPUP00000010705 | 98.8806 | 98.5075 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 99.6269 | 99.2537 | aotus_nancymaae | ENSANAP00000006160 | 99.6269 | 99.2537 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | saimiri_boliviensis_boliviensis | ENSSBOP00000039492 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | chlorocebus_sabaeus | ENSCSAP00000010903 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 96.2687 | mesocricetus_auratus | ENSMAUP00000006050 | 98.5075 | 96.2687 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 97.3881 | 96.2687 | phocoena_sinus | ENSPSNP00000024675 | 97.3881 | 96.2687 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 89.9254 | 87.3134 | camelus_dromedarius | ENSCDRP00005028519 | 80.602 | 78.2609 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 88.4328 | 86.9403 | carlito_syrichta | ENSTSYP00000032209 | 88.4328 | 86.9403 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 92.9104 | 89.9254 | panthera_tigris_altaica | ENSPTIP00000005332 | 78.7975 | 76.2658 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.8806 | 96.2687 | microtus_ochrogaster | ENSMOCP00000006897 | 98.8806 | 96.2687 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 64.5522 | 63.0597 | sorex_araneus | ENSSARP00000002840 | 96.1111 | 93.8889 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 96.2687 | peromyscus_maniculatus_bairdii | ENSPEMP00000010926 | 98.5075 | 96.2687 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 95.8955 | 93.6567 | bos_mutus | ENSBMUP00000014902 | 95.8955 | 93.6567 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.8806 | 96.6418 | cricetulus_griseus_chok1gshd | ENSCGRP00001012219 | 98.8806 | 96.6418 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 97.3881 | pteropus_vampyrus | ENSPVAP00000008911 | 98.5075 | 97.3881 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.1343 | 97.3881 | myotis_lucifugus | ENSMLUP00000000992 | 98.1343 | 97.3881 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 97.3881 | 95.1493 | vombatus_ursinus | ENSVURP00010006631 | 97.3881 | 95.1493 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 97.3881 | 95.1493 | vombatus_ursinus | ENSVURP00010002844 | 97.3881 | 95.1493 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.8806 | 98.5075 | neovison_vison | ENSNVIP00000003620 | 98.8806 | 98.5075 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 95.8955 | 93.6567 | bos_taurus | ENSBTAP00000017600 | 95.8955 | 93.6567 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | rhinopithecus_bieti | ENSRBIP00000034934 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.1343 | 97.7612 | canis_lupus_dingo | ENSCAFP00020019336 | 98.1343 | 97.7612 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 97.7612 | sciurus_vulgaris | ENSSVLP00005008854 | 98.5075 | 97.7612 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 97.3881 | 95.1493 | phascolarctos_cinereus | ENSPCIP00000033937 | 97.3881 | 95.1493 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.8806 | 97.0149 | chinchilla_lanigera | ENSCLAP00000017186 | 98.8806 | 97.0149 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 89.1791 | 86.194 | cervus_hanglu_yarkandensis | ENSCHYP00000030500 | 86.5942 | 83.6957 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 96.2687 | 94.0299 | catagonus_wagneri | ENSCWAP00000011365 | 96.2687 | 94.0299 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 97.3881 | ictidomys_tridecemlineatus | ENSSTOP00000020518 | 98.5075 | 97.3881 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | cebus_imitator | ENSCCAP00000012634 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 97.3881 | urocitellus_parryii | ENSUPAP00010004199 | 98.5075 | 97.3881 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 95.5224 | 93.2836 | moschus_moschiferus | ENSMMSP00000002208 | 95.5224 | 93.2836 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 100 | 100 | rhinopithecus_roxellana | ENSRROP00000016245 | 100 | 100 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.5075 | 96.2687 | mus_pahari | MGP_PahariEiJ_P0027890 | 98.1413 | 95.9108 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 98.1343 | 97.7612 | propithecus_coquereli | ENSPCOP00000008712 | 98.1343 | 97.7612 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 60.8209 | 47.7612 | drosophila_melanogaster | FBpp0080299 | 29.7445 | 23.3577 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 51.4925 | 40.2985 | ciona_intestinalis | ENSCINP00000018113 | 23.6301 | 18.4932 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 91.4179 | 80.9701 | callorhinchus_milii | ENSCMIP00000013045 | 86.5724 | 76.6784 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 95.1493 | 86.5672 | lepisosteus_oculatus | ENSLOCP00000007240 | 95.1493 | 86.5672 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 71.6418 | 62.6866 | ictalurus_punctatus | ENSIPUP00000000148 | 68.8172 | 60.2151 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 88.806 | 80.2239 | esox_lucius | ENSELUP00000022874 | 72.561 | 65.5488 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 90.2985 | 81.3433 | oncorhynchus_kisutch | ENSOKIP00005028775 | 93.0769 | 83.8462 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 89.1791 | 80.9701 | oncorhynchus_kisutch | ENSOKIP00005099091 | 91.9231 | 83.4615 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 89.1791 | 80.9701 | oncorhynchus_mykiss | ENSOMYP00000054177 | 91.9231 | 83.4615 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 90.2985 | 81.3433 | oncorhynchus_mykiss | ENSOMYP00000091480 | 93.0769 | 83.8462 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 90.2985 | 81.3433 | salmo_salar | ENSSSAP00000090897 | 93.0769 | 83.8462 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 89.1791 | 80.9701 | salmo_salar | ENSSSAP00000137484 | 91.9231 | 83.4615 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 89.1791 | 80.9701 | salmo_trutta | ENSSTUP00000017625 | 91.9231 | 83.4615 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 89.9254 | 80.9701 | salmo_trutta | ENSSTUP00000114086 | 92.6923 | 83.4615 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 82.0896 | 72.7612 | poecilia_reticulata | ENSPREP00000012552 | 80.8824 | 71.6912 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 86.5672 | 78.7313 | cyclopterus_lumpus | ENSCLMP00005050201 | 90.9804 | 82.7451 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 92.9104 | 88.806 | xenopus_tropicalis | ENSXETP00000051787 | 93.609 | 89.4737 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 96.2687 | 93.6567 | chrysemys_picta_bellii | ENSCPBP00000028710 | 96.2687 | 93.6567 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 95.1493 | 92.9104 | sphenodon_punctatus | ENSSPUP00000007015 | 95.1493 | 92.9104 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 59.3284 | 52.6119 | crocodylus_porosus | ENSCPRP00005006439 | 59.5506 | 52.809 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 81.3433 | 79.1045 | laticauda_laticaudata | ENSLLTP00000013040 | 94.7826 | 92.1739 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 95.1493 | 92.5373 | podarcis_muralis | ENSPMRP00000006957 | 95.1493 | 92.5373 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 94.0299 | 91.4179 | geospiza_fortis | ENSGFOP00000003589 | 96.1832 | 93.5115 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 94.403 | 85.8209 | erpetoichthys_calabaricus | ENSECRP00000006611 | 96.1977 | 87.4525 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 79.8507 | 72.3881 | dicentrarchus_labrax | ENSDLAP00005061925 | 91.453 | 82.906 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 96.2687 | 93.2836 | pelodiscus_sinensis | ENSPSIP00000018448 | 96.2687 | 93.2836 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 96.2687 | 93.6567 | gopherus_evgoodei | ENSGEVP00005017362 | 96.2687 | 93.6567 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 96.2687 | 93.6567 | chelonoidis_abingdonii | ENSCABP00000019549 | 96.2687 | 93.6567 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 96.2687 | 93.6567 | terrapene_carolina_triunguis | ENSTMTP00000017517 | 96.2687 | 93.6567 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 89.1791 | 80.9701 | hucho_hucho | ENSHHUP00000044583 | 91.9231 | 83.4615 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 43.6567 | 34.7015 | ciona_savignyi | ENSCSAVP00000016701 | 60.6218 | 48.1865 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 90.2985 | 81.3433 | oncorhynchus_tshawytscha | ENSOTSP00005062872 | 93.0769 | 83.8462 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 88.806 | 80.597 | oncorhynchus_tshawytscha | ENSOTSP00005042565 | 91.5385 | 83.0769 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 88.4328 | 80.2239 | nothobranchius_furzeri | ENSNFUP00015033284 | 91.1538 | 82.6923 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 73.1343 | 65.2985 | denticeps_clupeoides | ENSDCDP00000018197 | 80.9917 | 72.314 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 83.5821 | 73.5075 | larimichthys_crocea | ENSLCRP00005000831 | 80.5755 | 70.8633 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 91.791 | 84.3284 | paramormyrops_kingsleyae | ENSPKIP00000015593 | 91.791 | 84.3284 |
| ENSG00000019549 | homo_sapiens | ENSP00000020945 | 78.7313 | 66.791 | scleropages_formosus | ENSSFOP00015028084 | 77.0073 | 65.3285 |
Gene Ontology
| Go ID | Go term | No. evidence | Entries | Species | Category |
|---|---|---|---|---|---|
| GO:0000122 | involved_in negative regulation of transcription by RNA polymerase II | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:0000785 | part_of chromatin | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0000977 | enables RNA polymerase II transcription regulatory region sequence-specific DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0000978 | enables RNA polymerase II cis-regulatory region sequence-specific DNA binding | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0000981 | enables DNA-binding transcription factor activity, RNA polymerase II-specific | 1 | IBA | Homo_sapiens(9606) | Function |
| GO:0001227 | enables DNA-binding transcription repressor activity, RNA polymerase II-specific | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0001649 | involved_in osteoblast differentiation | 1 | IEP | Homo_sapiens(9606) | Process |
| GO:0001837 | involved_in epithelial to mesenchymal transition | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0003180 | involved_in aortic valve morphogenesis | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0003198 | involved_in epithelial to mesenchymal transition involved in endocardial cushion formation | 2 | ISS,TAS | Homo_sapiens(9606) | Process |
| GO:0003273 | involved_in cell migration involved in endocardial cushion formation | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0003682 | enables chromatin binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0005515 | enables protein binding | 1 | IPI | Homo_sapiens(9606) | Function |
| GO:0005634 | located_in nucleus | 1 | IDA | Homo_sapiens(9606) | Component |
| GO:0005654 | located_in nucleoplasm | 1 | TAS | Homo_sapiens(9606) | Component |
| GO:0005737 | located_in cytoplasm | 1 | IEA | Homo_sapiens(9606) | Component |
| GO:0006325 | involved_in chromatin organization | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0006355 | involved_in regulation of DNA-templated transcription | 1 | IBA | Homo_sapiens(9606) | Process |
| GO:0006933 | involved_in negative regulation of cell adhesion involved in substrate-bound cell migration | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0007219 | involved_in Notch signaling pathway | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0007605 | involved_in sensory perception of sound | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0010839 | involved_in negative regulation of keratinocyte proliferation | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0010957 | involved_in negative regulation of vitamin D biosynthetic process | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0014032 | involved_in neural crest cell development | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0030335 | involved_in positive regulation of cell migration | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032331 | involved_in negative regulation of chondrocyte differentiation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0032642 | involved_in regulation of chemokine production | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0033028 | involved_in myeloid cell apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0033033 | involved_in negative regulation of myeloid cell apoptotic process | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0033629 | acts_upstream_of negative regulation of cell adhesion mediated by integrin | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0035921 | involved_in desmosome disassembly | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0043473 | involved_in pigmentation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0043518 | involved_in negative regulation of DNA damage response, signal transduction by p53 class mediator | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0043542 | involved_in endothelial cell migration | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0043565 | enables sequence-specific DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0045600 | involved_in positive regulation of fat cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0045667 | involved_in regulation of osteoblast differentiation | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:0046872 | enables metal ion binding | 1 | IEA | Homo_sapiens(9606) | Function |
| GO:0050872 | involved_in white fat cell differentiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060021 | involved_in roof of mouth development | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060429 | involved_in epithelium development | 1 | ISS | Homo_sapiens(9606) | Process |
| GO:0060536 | involved_in cartilage morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0060693 | involved_in regulation of branching involved in salivary gland morphogenesis | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0061314 | involved_in Notch signaling involved in heart development | 1 | TAS | Homo_sapiens(9606) | Process |
| GO:0070563 | involved_in negative regulation of vitamin D receptor signaling pathway | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0070888 | enables E-box binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:0071364 | involved_in cellular response to epidermal growth factor stimulus | 1 | IDA | Homo_sapiens(9606) | Process |
| GO:0071425 | involved_in hematopoietic stem cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0071479 | involved_in cellular response to ionizing radiation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:0090090 | involved_in negative regulation of canonical Wnt signaling pathway | 2 | IDA,IMP | Homo_sapiens(9606) | Process |
| GO:1902034 | involved_in negative regulation of hematopoietic stem cell proliferation | 1 | IEA | Homo_sapiens(9606) | Process |
| GO:1902230 | involved_in negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:1990837 | enables sequence-specific double-stranded DNA binding | 1 | IDA | Homo_sapiens(9606) | Function |
| GO:2000810 | involved_in regulation of bicellular tight junction assembly | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:2000811 | involved_in negative regulation of anoikis | 1 | IMP | Homo_sapiens(9606) | Process |
| GO:2001240 | involved_in negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1 | ISS | Homo_sapiens(9606) | Process |