| RBP Type |
N.A. |
| Diseases |
|
| Drug |
|
| Main interacting RNAs |
|
| Moonlighting functions |
|
| Localizations |
|
|
BulkPerturb-seq
|
|
| Ensembl ID | ENSCHAG00000024918 | Gene ID |
|
Accession |
- |
| Symbol |
helz2 |
Alias |
- |
Full Name |
helicase with zinc finger 2, transcriptional coactivator |
| Status |
Confidence |
Length |
20510 bases |
Strand |
Plus strand |
| Position |
5 : 20344287 - 20364796 |
RNA binding domain |
OB_Dis3 , RNB |
| Summary |
N.A. |
RNA binding domains (RBDs)

| Protein |
Domain |
Pfam ID |
E-value |
Domain number |
| ENSCHAP00000055003 |
OB_Dis3 |
PF17849 |
1.4e-05 |
1 |
| ENSCHAP00000055003 |
RNB |
PF00773 |
2.2e-58 |
2 |
| ENSCHAP00000054985 |
OB_Dis3 |
PF17849 |
1.5e-05 |
1 |
| ENSCHAP00000054985 |
RNB |
PF00773 |
3.9e-57 |
2 |
RNA binding proteomes (RBPomes)
| Pubmed ID |
Full Name |
Cell |
Author |
Time |
Doi |
Literatures on RNA binding capacity
| Pubmed ID |
Title |
Author |
Time |
Journal |
| Name |
Transcript ID |
bp |
Protein |
Translation ID |
| HELZ2-201 |
ENSCHAT00000059298 |
10171 |
2875aa |
ENSCHAP00000054985 |
| HELZ2-202 |
ENSCHAT00000059317 |
9667 |
2707aa |
ENSCHAP00000055003 |
| Pathway ID |
Pathway Name |
Source |
| | |
| ensgID |
Trait |
pValue |
Pubmed ID |
| ensgID | SNP | Chromosome | Position |
Trait | PubmedID | Or or BEAT |
EFO ID |
|---|
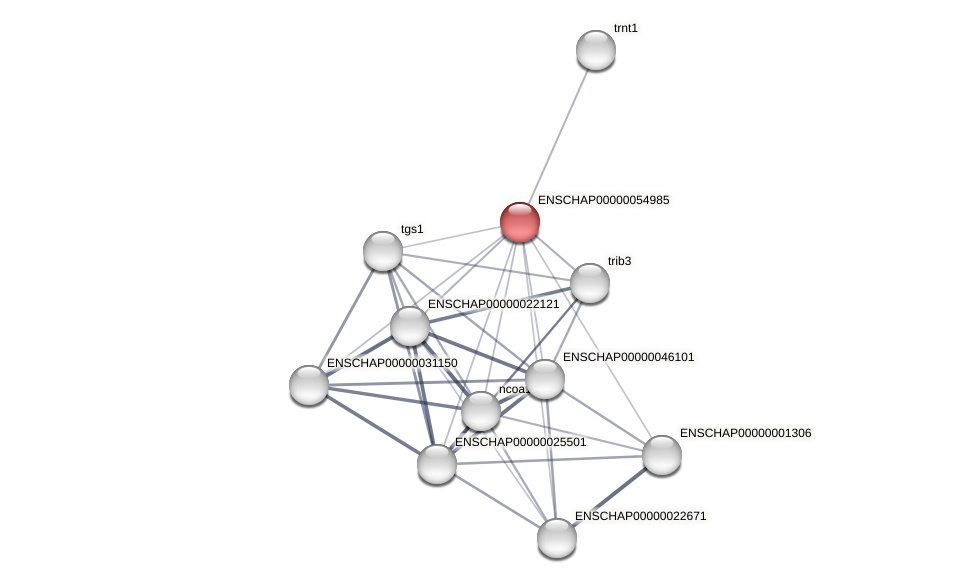
Protein-Protein Interaction (PPI)
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000004762 | 27.2911 | 14.0836 | clupea_harengus | ENSCHAP00000054985 | 14.087 | 7.26957 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000057740 | 22.7925 | 11.0918 | clupea_harengus | ENSCHAP00000054985 | 18.2261 | 8.86957 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000043629 | 30.384 | 16.778 | clupea_harengus | ENSCHAP00000054985 | 12.6609 | 6.9913 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000044540 | 25.2465 | 14.3984 | clupea_harengus | ENSCHAP00000054985 | 8.90435 | 5.07826 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000053122 | 35.3316 | 20.0255 | clupea_harengus | ENSCHAP00000054985 | 9.63478 | 5.46087 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000056276 | 18.7566 | 10.2213 | clupea_harengus | ENSCHAP00000054985 | 12.3826 | 6.74783 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000055028 | 59.7458 | 43.4746 | clupea_harengus | ENSCHAP00000054985 | 49.0435 | 35.687 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000020927 | 19.4005 | 10.0817 | clupea_harengus | ENSCHAP00000054985 | 12.3826 | 6.43478 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000025229 | 37.8038 | 22.5923 | clupea_harengus | ENSCHAP00000054985 | 14.6087 | 8.73043 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000040423 | 37.646 | 22.1923 | clupea_harengus | ENSCHAP00000054985 | 14.5739 | 8.5913 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000023655 | 32.1074 | 17.1968 | clupea_harengus | ENSCHAP00000054985 | 11.2348 | 6.01739 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000010459 | 31.2312 | 18.1181 | clupea_harengus | ENSCHAP00000054985 | 10.8522 | 6.29565 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000024849 | 69.869 | 58.0786 | clupea_harengus | ENSCHAP00000054985 | 5.56522 | 4.62609 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000024951 | 60.5941 | 42.5743 | clupea_harengus | ENSCHAP00000054985 | 10.6435 | 7.47826 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000055016 | 67.1053 | 51.7544 | clupea_harengus | ENSCHAP00000054985 | 5.32174 | 4.10435 |
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 21.8087 | 11.4435 | caenorhabditis_elegans | ZK1067.2b.1 | 25.6652 | 13.467 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 22.7478 | 11.6174 | drosophila_melanogaster | FBpp0271875 | 39.0215 | 19.9284 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 4.10435 | 2.50435 | ciona_intestinalis | ENSCINP00000031582 | 56.1905 | 34.2857 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 5.32174 | 4.06957 | ciona_intestinalis | ENSCINP00000019982 | 53.8732 | 41.1972 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 7.06087 | 4.97391 | ciona_intestinalis | ENSCINP00000016680 | 53.1414 | 37.4346 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 11.1304 | 8.52174 | petromyzon_marinus | ENSPMAP00000008627 | 66.39 | 50.8299 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 60.1739 | 42.8522 | callorhinchus_milii | ENSCMIP00000030536 | 61.1956 | 43.5798 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 59.1304 | 42.087 | latimeria_chalumnae | ENSLACP00000009242 | 58.9664 | 41.9702 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 66.6087 | 51.2 | lepisosteus_oculatus | ENSLOCP00000002057 | 67.2166 | 51.6673 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 59.6174 | 43.6522 | gallus_gallus | ENSGALP00010022854 | 60.8233 | 44.5351 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 37.287 | 25.3565 | erinaceus_europaeus | ENSEEUP00000004290 | 43.8984 | 29.8526 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 57.4957 | 41.0435 | sarcophilus_harrisii | ENSSHAP00000021431 | 57.2567 | 40.8729 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.8174 | 42.6435 | ornithorhynchus_anatinus | ENSOANP00000043689 | 59.1466 | 42.8821 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 25.1478 | 17.0087 | echinops_telfairi | ENSETEP00000008083 | 31.2716 | 21.1505 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 13.0087 | 10.5043 | electrophorus_electricus | ENSEEEP00000017870 | 69.6462 | 56.2384 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 6.15652 | 5.25217 | electrophorus_electricus | ENSEEEP00000017868 | 80.0905 | 68.3258 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 69.7043 | 54.9217 | astyanax_mexicanus | ENSAMXP00000017981 | 71.2914 | 56.1722 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 51.1652 | 36 | gorilla_gorilla | ENSGGOP00000008453 | 55.4467 | 39.0124 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 51.0609 | 35.8957 | panthera_pardus | ENSPPRP00000021841 | 55.7963 | 39.2246 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 51.4087 | 36.2783 | notamacropus_eugenii | ENSMEUP00000005653 | 58.0063 | 40.9341 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.1652 | 38.5739 | mus_caroli | MGP_CAROLIEiJ_P0056689 | 53.7992 | 37.6187 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 53.913 | 37.913 | macaca_nemestrina | ENSMNEP00000044497 | 54.4433 | 38.2859 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.5826 | 39.0261 | ailuropoda_melanoleuca | ENSAMEP00000022463 | 52.3591 | 36.7628 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 5.42609 | 4.03478 | canis_lupus_familiaris | ENSCAFP00845030551 | 62.4 | 46.4 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.8174 | 38.713 | felis_catus | ENSFCAP00000056738 | 53.8251 | 38.0123 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 70.0522 | 53.9826 | esox_lucius | ENSELUP00000013163 | 71.7492 | 55.2903 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 53.7739 | 37.8087 | cercocebus_atys | ENSCATP00000003767 | 54.2647 | 38.1537 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.7826 | 42.1565 | parus_major | ENSPMJP00000008717 | 59.6751 | 42.7966 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.1304 | 38.6783 | mus_pahari | MGP_PahariEiJ_P0067714 | 53.6924 | 37.6694 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.6435 | 38.6435 | panthera_leo | ENSPLOP00000024794 | 53.6727 | 37.957 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 41.5304 | 29.1826 | procavia_capensis | ENSPCAP00000001064 | 54.9977 | 38.6458 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 51.2 | 35.7217 | pan_troglodytes | ENSPTRP00000023668 | 55.1724 | 38.4933 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 69.0435 | 53.9826 | salmo_salar | ENSSSAP00000097103 | 73.2202 | 57.2482 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 47.7565 | 33.0783 | ursus_americanus | ENSUAMP00000033090 | 53.0731 | 36.7607 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 5.04348 | 3.61739 | ursus_americanus | ENSUAMP00000033087 | 68.0751 | 48.8263 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 69.0087 | 53.8435 | oncorhynchus_kisutch | ENSOKIP00005100090 | 73.0486 | 56.9956 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.887 | 42.6087 | serinus_canaria | ENSSCAP00000019989 | 60.0568 | 43.4551 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 41.4957 | 29.6 | pseudonaja_textilis | ENSPTXP00000018941 | 61.3999 | 43.7982 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 46.2609 | 32.313 | dasypus_novemcinctus | ENSDNOP00000013821 | 53.0938 | 37.0858 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 49.3217 | 34.4 | mandrillus_leucophaeus | ENSMLEP00000020335 | 54.2256 | 37.8203 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.0261 | 38.5739 | rattus_norvegicus | ENSRNOP00000017787 | 53.3199 | 37.3778 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 30.8174 | 20.9043 | sorex_araneus | ENSSARP00000009275 | 38.5385 | 26.1418 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 59.3391 | 42.887 | meleagris_gallopavo | ENSMGAP00000007028 | 60.368 | 43.6306 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.2609 | 37.4957 | xenopus_tropicalis | ENSXETP00000057738 | 56.6038 | 39.1147 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 53.0087 | 36.2087 | sus_scrofa | ENSSSCP00000003585 | 52.5155 | 35.8718 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 51.2 | 35.8261 | pan_paniscus | ENSPPAP00000029228 | 55.5681 | 38.8826 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.8522 | 37.6348 | otolemur_garnettii | ENSOGAP00000021455 | 53.7308 | 36.8654 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.8522 | 38.5739 | mustela_putorius_furo | ENSMPUP00000008298 | 53.6942 | 37.7596 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 59.0609 | 43.5478 | chrysemys_picta_bellii | ENSCPBP00000020341 | 60.5132 | 44.6187 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.1913 | 41.2522 | laticauda_laticaudata | ENSLLTP00000006016 | 59.7713 | 42.3723 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 9.0087 | 6.50435 | geospiza_fortis | ENSGFOP00000001218 | 49.9037 | 36.0308 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 6.1913 | 5.0087 | geospiza_fortis | ENSGFOP00000001204 | 72.9508 | 59.0164 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 23.2 | 16.5565 | sphenodon_punctatus | ENSSPUP00000024244 | 61.418 | 43.8306 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.2609 | 37.287 | dipodomys_ordii | ENSDORP00000005236 | 54.5264 | 37.4694 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.2609 | 37.913 | mus_spretus | MGP_SPRETEiJ_P0058920 | 53.7375 | 37.5474 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 59.2 | 42.713 | anas_platyrhynchos_platyrhynchos | ENSAPLP00000010126 | 60.3118 | 43.5152 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.2696 | 38.3304 | vulpes_vulpes | ENSVVUP00000020600 | 53.0905 | 36.8192 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.3391 | 38.887 | cricetulus_griseus_chok1gshd | ENSCGRP00001016417 | 54.7865 | 38.4986 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.087 | 41.6348 | phascolarctos_cinereus | ENSPCIP00000023729 | 58.0063 | 41.5769 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.1652 | 38.2261 | rhinolophus_ferrumequinum | ENSRFEP00010019600 | 53.6899 | 37.2038 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.8609 | 39.0609 | myotis_lucifugus | ENSMLUP00000013287 | 54.9248 | 38.4063 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 65.0087 | 48.8696 | danio_rerio | ENSDARP00000128833 | 70.369 | 52.8991 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.2696 | 38.4 | mus_musculus | ENSMUSP00000091756 | 53.5017 | 37.1717 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.7565 | 38.6435 | equus_caballus | ENSECAP00000079324 | 53.63 | 37.1696 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.713 | 42.9565 | anolis_carolinensis | ENSACAP00000040165 | 59.7522 | 43.7168 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 53.8435 | 37.7391 | papio_anubis | ENSPANP00000026542 | 54.8547 | 38.4479 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 69.4957 | 53.913 | salmo_trutta | ENSSTUP00000070935 | 73.1868 | 56.7766 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.0261 | 38.6087 | jaculus_jaculus | ENSJJAP00000021420 | 54.3269 | 38.1181 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 61.913 | 45.1826 | erpetoichthys_calabaricus | ENSECRP00000016982 | 62.3031 | 45.4673 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.5043 | 42.4 | podarcis_muralis | ENSPMRP00000006663 | 59.5398 | 43.1504 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 5.56522 | 4.66087 | chelonoidis_abingdonii | ENSCABP00000000276 | 69.869 | 58.5153 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 57.9826 | 41.913 | crocodylus_porosus | ENSCPRP00005009669 | 57.5026 | 41.5661 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 53.7739 | 37.7739 | rhinopithecus_roxellana | ENSRROP00000002276 | 54.7838 | 38.4833 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 57.9478 | 41.3913 | vombatus_ursinus | ENSVURP00010019379 | 57.8673 | 41.3338 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 40.3826 | 28.2783 | sciurus_vulgaris | ENSSVLP00005014012 | 56.3592 | 39.466 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.3652 | 41.7043 | monodelphis_domestica | ENSMODP00000021169 | 58.4873 | 41.7916 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 53.913 | 38.0174 | macaca_mulatta | ENSMMUP00000051269 | 54.9256 | 38.7314 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.887 | 40.4522 | pelodiscus_sinensis | ENSPSIP00000014146 | 57.2777 | 42.2142 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 42.6435 | 30.087 | notechis_scutatus | ENSNSUP00000024379 | 61.2388 | 43.2068 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 45.2522 | 31.5826 | pteropus_vampyrus | ENSPVAP00000006780 | 52.6934 | 36.776 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.5043 | 38.1565 | ictidomys_tridecemlineatus | ENSSTOP00000015858 | 54.3908 | 38.0771 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 13.6 | 10.3304 | carassius_auratus | ENSCARP00000044144 | 70.4504 | 53.5135 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 6.53913 | 5.28696 | carassius_auratus | ENSCARP00000085241 | 81.0345 | 65.5172 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 50.4348 | 37.8783 | carassius_auratus | ENSCARP00000085253 | 71.8533 | 53.9643 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 63.1652 | 48.6609 | carassius_auratus | ENSCARP00000008579 | 71.4679 | 55.0571 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 6.9913 | 5.70435 | carassius_auratus | ENSCARP00000044113 | 81.0484 | 66.129 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 5.73913 | 4.34783 | panthera_tigris_altaica | ENSPTIP00000021122 | 71.7391 | 54.3478 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.1217 | 38.087 | marmota_marmota_marmota | ENSMMMP00000002058 | 53.2148 | 37.4487 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 50.8522 | 35.5478 | aotus_nancymaae | ENSANAP00000003267 | 55.0659 | 38.4934 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 65.4609 | 50.5391 | pygocentrus_nattereri | ENSPNAP00000030210 | 74.3874 | 57.4308 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.513 | 38.9565 | octodon_degus | ENSODEP00000009626 | 54.8831 | 38.5144 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.9217 | 42.7826 | ficedula_albicollis | ENSFALP00000006090 | 59.7742 | 43.4016 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 10.8174 | 7.44348 | nomascus_leucogenys | ENSNLEP00000038405 | 48.5179 | 33.3853 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 50.2261 | 35.2 | nomascus_leucogenys | ENSNLEP00000012302 | 55.2833 | 38.7443 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 49.0435 | 33.9826 | microcebus_murinus | ENSMICP00000038874 | 53.3687 | 36.9796 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.8174 | 42.9565 | aquila_chrysaetos_chrysaetos | ENSACCP00020000847 | 58.5526 | 42.7632 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 50.7826 | 35.4087 | saimiri_boliviensis_boliviensis | ENSSBOP00000034324 | 55.0736 | 38.4006 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.087 | 38.3652 | neovison_vison | ENSNVIP00000025100 | 55.0248 | 39.0304 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 4.8 | 3.65217 | catagonus_wagneri | ENSCWAP00000010730 | 48.4211 | 36.8421 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 3.89565 | 2.78261 | phocoena_sinus | ENSPSNP00000026687 | 36.3636 | 25.974 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 13.6348 | 10.1913 | loxodonta_africana | ENSLAFP00000005372 | 60.2151 | 45.0077 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 68.7652 | 53.6348 | hucho_hucho | ENSHHUP00000013437 | 72.8714 | 56.8374 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.713 | 42.9217 | taeniopygia_guttata | ENSTGUP00000007193 | 59.8582 | 43.7589 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.1304 | 38.6087 | mesocricetus_auratus | ENSMAUP00000001164 | 54.674 | 38.2891 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.087 | 42.5739 | strigops_habroptila | ENSSHBP00005014903 | 59.136 | 43.3428 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 49.9478 | 34.4696 | carlito_syrichta | ENSTSYP00000017574 | 56.6693 | 39.1081 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 65.5652 | 52.487 | denticeps_clupeoides | ENSDCDP00000021237 | 75.6117 | 60.5295 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.2 | 38.5739 | mus_spicilegus | ENSMSIP00000015521 | 53.8514 | 37.6315 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.6783 | 38.9217 | cavia_porcellus | ENSCPOP00000006228 | 53.433 | 38.0354 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 51.1304 | 35.8261 | cebus_imitator | ENSCCAP00000019826 | 55.5976 | 38.9561 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.8261 | 39.4435 | chinchilla_lanigera | ENSCLAP00000009008 | 55.2686 | 39.0496 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.1217 | 37.9478 | urocitellus_parryii | ENSUPAP00010005658 | 53.4157 | 37.4528 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 65.1826 | 49.2522 | paramormyrops_kingsleyae | ENSPKIP00000021724 | 68.9478 | 52.0971 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 59.1304 | 43.7565 | gopherus_evgoodei | ENSGEVP00005014173 | 60.1557 | 44.5152 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 59.1652 | 43.2696 | struthio_camelus_australis | ENSSCUP00000017277 | 60.3191 | 44.1135 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 53.7391 | 37.7739 | chlorocebus_sabaeus | ENSCSAP00000012483 | 54.7484 | 38.4833 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 59.0609 | 43.5826 | terrapene_carolina_triunguis | ENSTMTP00000000888 | 60.1488 | 44.3854 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 68.9739 | 53.8435 | oncorhynchus_tshawytscha | ENSOTSP00005033880 | 73.0118 | 56.9956 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 59.8957 | 43.7217 | coturnix_japonica | ENSCJPP00005006285 | 61.1289 | 44.6219 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 59.4783 | 43.4087 | anser_brachyrhynchus | ENSABRP00000008068 | 60.5739 | 44.2083 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 65.7043 | 50.1913 | cyprinus_carpio_carpio | ENSCCRP00000133767 | 70.8021 | 54.0855 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 65.1478 | 49.7739 | cyprinus_carpio_carpio | ENSCCRP00000136574 | 70.5196 | 53.878 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 63.4435 | 48.4174 | cyprinus_carpio_carpio | ENSCCRP00000144388 | 71.8676 | 54.8463 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.5391 | 38.087 | peromyscus_maniculatus_bairdii | ENSPEMP00000017742 | 55.1142 | 38.4886 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.4435 | 39.2696 | nannospalax_galili | ENSNGAP00000011802 | 54.3471 | 38.493 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 51.1304 | 35.6522 | rhinopithecus_bieti | ENSRBIP00000042854 | 55.4926 | 38.6938 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 51.3043 | 35.9304 | homo_sapiens | ENSP00000417401 | 55.6814 | 38.9958 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 58.4696 | 41.913 | salvator_merianae | ENSSMRP00000025183 | 59.7158 | 42.8064 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 6.74783 | 5.42609 | sinocyclocheilus_grahami | ENSSGRP00000020412 | 81.1715 | 65.272 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 49.7391 | 38.2609 | sinocyclocheilus_grahami | ENSSGRP00000020415 | 73.8636 | 56.8182 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 54.7478 | 37.913 | leptobrachium_leishanense | ENSLLEP00000015739 | 56.7412 | 39.2934 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 69.5652 | 54.1217 | oncorhynchus_mykiss | ENSOMYP00000031172 | 71.7875 | 55.8507 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 55.8609 | 38.6783 | microtus_ochrogaster | ENSMOCP00000017809 | 54.2751 | 37.5803 |
| ENSCHAG00000024918 | clupea_harengus | ENSCHAP00000054985 | 49.8087 | 34.2957 | propithecus_coquereli | ENSPCOP00000011268 | 54.8659 | 37.7778 |
| Go ID |
Go term |
No. evidence |
Entries |
Species |
Category |