| RBP Type |
N.A. |
| Diseases |
|
| Drug |
|
| Main interacting RNAs |
|
| Moonlighting functions |
|
| Localizations |
|
|
BulkPerturb-seq
|
|
| Ensembl ID | ENSBSLG00000001086 | Gene ID |
114849329 |
Accession |
- |
| Symbol |
eif4a1b |
Alias |
- |
Full Name |
eukaryotic initiation factor 4A-I |
| Status |
Confidence |
Length |
4815 bases |
Strand |
Plus strand |
| Position |
2 : 16153616 - 16158430 |
RNA binding domain |
ResIII , DEAD |
| Summary |
N.A. |
RNA binding domains (RBDs)

| Protein |
Domain |
Pfam ID |
E-value |
Domain number |
| ENSBSLP00000001918 |
ResIII |
PF04851 |
6.6e-07 |
1 |
| ENSBSLP00000001918 |
DEAD |
PF00270 |
1.8e-44 |
2 |
| ENSBSLP00000001921 |
ResIII |
PF04851 |
6.9e-07 |
1 |
| ENSBSLP00000001921 |
DEAD |
PF00270 |
1.8e-44 |
2 |
| ENSBSLP00000001917 |
ResIII |
PF04851 |
7e-07 |
1 |
| ENSBSLP00000001917 |
DEAD |
PF00270 |
1.8e-44 |
2 |
| ENSBSLP00000001911 |
ResIII |
PF04851 |
7e-07 |
1 |
| ENSBSLP00000001911 |
DEAD |
PF00270 |
1.8e-44 |
2 |
| ENSBSLP00000001923 |
ResIII |
PF04851 |
7e-07 |
1 |
| ENSBSLP00000001923 |
DEAD |
PF00270 |
1.8e-44 |
2 |
| ENSBSLP00000001914 |
ResIII |
PF04851 |
7.1e-07 |
1 |
| ENSBSLP00000001914 |
DEAD |
PF00270 |
1.8e-44 |
2 |
| ENSBSLP00000001912 |
ResIII |
PF04851 |
7.2e-07 |
1 |
| ENSBSLP00000001912 |
DEAD |
PF00270 |
1.9e-44 |
2 |
| ENSBSLP00000001916 |
ResIII |
PF04851 |
7.3e-07 |
1 |
| ENSBSLP00000001916 |
DEAD |
PF00270 |
2e-44 |
2 |
| ENSBSLP00000001919 |
ResIII |
PF04851 |
7.6e-07 |
1 |
| ENSBSLP00000001919 |
DEAD |
PF00270 |
1.8e-44 |
2 |
| ENSBSLP00000001920 |
ResIII |
PF04851 |
7.9e-07 |
1 |
| ENSBSLP00000001920 |
DEAD |
PF00270 |
2.1e-44 |
2 |
RNA binding proteomes (RBPomes)
| Pubmed ID |
Full Name |
Cell |
Author |
Time |
Doi |
Literatures on RNA binding capacity
| Pubmed ID |
Title |
Author |
Time |
Journal |
| Name |
Transcript ID |
bp |
Protein |
Translation ID |
| eif4a1a-201 |
ENSBSLT00000002263 |
1884 |
404aa |
ENSBSLP00000001911 |
| eif4a1a-202 |
ENSBSLT00000002264 |
1899 |
409aa |
ENSBSLP00000001912 |
| eif4a1a-203 |
ENSBSLT00000002266 |
1887 |
405aa |
ENSBSLP00000001914 |
| eif4a1a-204 |
ENSBSLT00000002268 |
1920 |
416aa |
ENSBSLP00000001916 |
| eif4a1a-205 |
ENSBSLT00000002269 |
1887 |
405aa |
ENSBSLP00000001917 |
| eif4a1a-206 |
ENSBSLT00000002270 |
1887 |
405aa |
ENSBSLP00000001918 |
| eif4a1a-207 |
ENSBSLT00000002271 |
1869 |
399aa |
ENSBSLP00000001919 |
| eif4a1a-208 |
ENSBSLT00000002272 |
1965 |
431aa |
ENSBSLP00000001920 |
| eif4a1a-209 |
ENSBSLT00000002273 |
1878 |
402aa |
ENSBSLP00000001921 |
| eif4a1a-210 |
ENSBSLT00000002275 |
1887 |
405aa |
ENSBSLP00000001923 |
| Pathway ID |
Pathway Name |
Source |
| | |
| ensgID |
Trait |
pValue |
Pubmed ID |
| ensgID | SNP | Chromosome | Position |
Trait | PubmedID | Or or BEAT |
EFO ID |
|---|
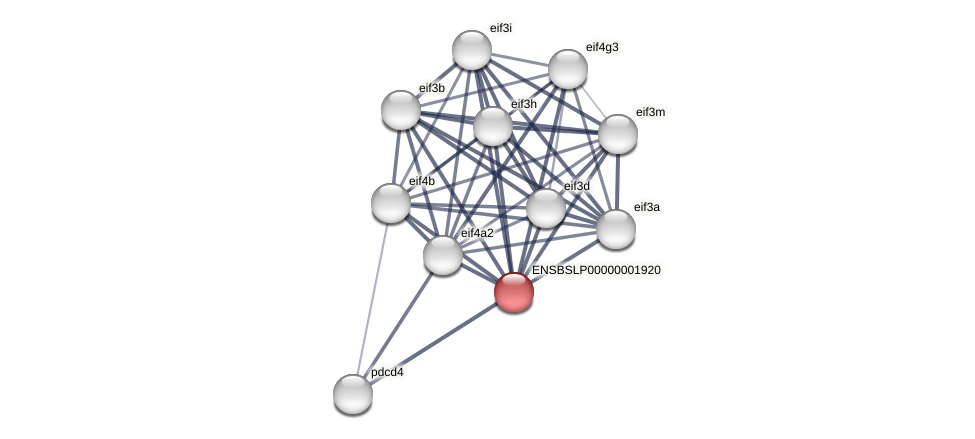
Protein-Protein Interaction (PPI)
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000050134 | 95.1338 | 87.5912 | betta_splendens | ENSBSLP00000001920 | 90.7193 | 83.5267 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000054819 | 28.2799 | 15.5977 | betta_splendens | ENSBSLP00000001920 | 45.0116 | 24.826 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000021766 | 36.8973 | 24.109 | betta_splendens | ENSBSLP00000001920 | 40.8353 | 26.6821 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000044061 | 31.7204 | 18.2796 | betta_splendens | ENSBSLP00000001920 | 41.0673 | 23.6659 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000022712 | 18.6941 | 10.9123 | betta_splendens | ENSBSLP00000001920 | 48.4919 | 28.3063 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000004272 | 15.0808 | 7.80969 | betta_splendens | ENSBSLP00000001920 | 38.9791 | 20.1856 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000000850 | 38.0252 | 24.1597 | betta_splendens | ENSBSLP00000001920 | 41.9954 | 26.6821 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000033887 | 26.9179 | 17.2275 | betta_splendens | ENSBSLP00000001920 | 46.4037 | 29.6984 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000050852 | 28.6285 | 17.0439 | betta_splendens | ENSBSLP00000001920 | 49.884 | 29.6984 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000022510 | 31.2796 | 18.1675 | betta_splendens | ENSBSLP00000001920 | 45.9397 | 26.6821 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000024951 | 22.9508 | 14.9883 | betta_splendens | ENSBSLP00000001920 | 45.4756 | 29.6984 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000024836 | 30.7692 | 18.3946 | betta_splendens | ENSBSLP00000001920 | 42.6914 | 25.522 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000020240 | 33.1699 | 22.0588 | betta_splendens | ENSBSLP00000001920 | 47.0998 | 31.3225 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001613 | 30.6306 | 20.7207 | betta_splendens | ENSBSLP00000001920 | 47.3318 | 32.0186 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000007016 | 31.1927 | 20.948 | betta_splendens | ENSBSLP00000001920 | 47.3318 | 31.7865 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000033120 | 31.4554 | 21.5962 | betta_splendens | ENSBSLP00000001920 | 46.6357 | 32.0186 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000026511 | 35.4955 | 19.8198 | betta_splendens | ENSBSLP00000001920 | 45.7077 | 25.522 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000034050 | 23.2727 | 13.3333 | betta_splendens | ENSBSLP00000001920 | 44.5476 | 25.522 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000017813 | 30.3859 | 18.1672 | betta_splendens | ENSBSLP00000001920 | 43.8515 | 26.2181 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000051302 | 42.9448 | 25.3579 | betta_splendens | ENSBSLP00000001920 | 48.7239 | 28.7703 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000005085 | 42.2222 | 25.2525 | betta_splendens | ENSBSLP00000001920 | 48.4919 | 29.0023 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000015775 | 73.5225 | 59.1017 | betta_splendens | ENSBSLP00000001920 | 72.1578 | 58.0046 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000032337 | 28.1294 | 17.5809 | betta_splendens | ENSBSLP00000001920 | 46.4037 | 29.0023 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000021719 | 27.5116 | 15.456 | betta_splendens | ENSBSLP00000001920 | 41.2993 | 23.2019 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000012893 | 30.6376 | 18.507 | betta_splendens | ENSBSLP00000001920 | 45.7077 | 27.6102 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000021539 | 27.2727 | 16.6453 | betta_splendens | ENSBSLP00000001920 | 49.42 | 30.1624 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000047919 | 28.5521 | 17.1854 | betta_splendens | ENSBSLP00000001920 | 48.9559 | 29.4664 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000027362 | 23.31 | 13.4033 | betta_splendens | ENSBSLP00000001920 | 46.4037 | 26.6821 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000024709 | 25 | 14.875 | betta_splendens | ENSBSLP00000001920 | 46.4037 | 27.6102 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000005424 | 20.9713 | 12.6932 | betta_splendens | ENSBSLP00000001920 | 44.0835 | 26.6821 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000015494 | 29.4562 | 16.3142 | betta_splendens | ENSBSLP00000001920 | 45.2436 | 25.058 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000038378 | 51.7454 | 34.9076 | betta_splendens | ENSBSLP00000001920 | 58.4687 | 39.4432 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000013458 | 51.6393 | 33.6066 | betta_splendens | ENSBSLP00000001920 | 58.4687 | 38.051 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000035664 | 24.4552 | 13.4383 | betta_splendens | ENSBSLP00000001920 | 46.8677 | 25.7541 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000012999 | 27.3842 | 15.8038 | betta_splendens | ENSBSLP00000001920 | 46.6357 | 26.9142 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000008417 | 28.8052 | 15.5483 | betta_splendens | ENSBSLP00000001920 | 40.8353 | 22.0418 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000057307 | 42.7105 | 25.6674 | betta_splendens | ENSBSLP00000001920 | 48.2599 | 29.0023 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000020855 | 49.5455 | 31.5909 | betta_splendens | ENSBSLP00000001920 | 50.58 | 32.2506 |
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.3434 | 90.7193 | fundulus_heteroclitus | ENSFHEP00000013320 | 87.0897 | 85.558 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.0232 | 88.8631 | poecilia_reticulata | ENSPREP00000005047 | 99.4872 | 98.2051 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.8793 | 90.4872 | xiphophorus_maculatus | ENSXMAP00000036772 | 98.0198 | 96.5347 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.6473 | 88.8631 | oryzias_latipes | ENSORLP00000013361 | 97.5309 | 94.5679 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.3271 | 86.0789 | cyclopterus_lumpus | ENSCLMP00005045223 | 91.4489 | 88.1235 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.3271 | 87.703 | oreochromis_niloticus | ENSONIP00000051226 | 91.4489 | 89.7862 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.3434 | 90.4872 | astatotilapia_calliptera | ENSACLP00000019156 | 98.5149 | 96.5347 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.7193 | 87.239 | sparus_aurata | ENSSAUP00010050924 | 95.1338 | 91.4842 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.3434 | 90.4872 | haplochromis_burtoni | ENSHBUP00000003584 | 98.5149 | 96.5347 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 90.2552 | lates_calcarifer | ENSLCAP00010026246 | 96.8293 | 94.8781 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.8793 | 90.2552 | cyprinodon_variegatus | ENSCVAP00000030564 | 98.0198 | 96.2871 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.6473 | 89.0951 | oryzias_melastigma | ENSOMEP00000002402 | 97.7723 | 95.0495 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.8793 | 89.5592 | kryptolebias_marmoratus | ENSKMAP00000009253 | 97.7778 | 95.3086 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.0232 | 86.5429 | dicentrarchus_labrax | ENSDLAP00005017536 | 86.031 | 82.7051 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 90.2552 | seriola_dumerili | ENSSDUP00000006690 | 98.2673 | 96.2871 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.4872 | 87.239 | labrus_bergylta | ENSLBEP00000024854 | 96.5347 | 93.0693 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 86.0789 | 82.8306 | scophthalmus_maximus | ENSSMAP00000057466 | 84.5103 | 81.3212 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.6473 | 90.0232 | poecilia_latipinna | ENSPLAP00000014923 | 97.5309 | 95.8025 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 30.6264 | 30.1624 | amphilophus_citrinellus | ENSACIP00000011489 | 100 | 98.4848 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 38.5151 | 36.6589 | amphilophus_citrinellus | ENSACIP00000019145 | 92.7374 | 88.2682 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 90.7193 | poecilia_formosa | ENSPFOP00000012514 | 98.2673 | 96.7822 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.6473 | 89.0951 | cottoperca_gobio | ENSCGOP00000034611 | 97.5309 | 94.8148 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.1833 | 89.3271 | acanthochromis_polyacanthus | ENSAPOP00000013720 | 97.2772 | 95.297 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 88.3991 | gasterosteus_aculeatus | ENSGACP00000026522 | 96.3592 | 92.4757 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 90.4872 | amphiprion_ocellaris | ENSAOCP00000024323 | 98.2673 | 96.5347 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.0232 | 85.6149 | hippocampus_comes | ENSHCOP00000019529 | 96.0396 | 91.3366 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.0951 | 85.3828 | sander_lucioperca | ENSSLUP00000028256 | 92.9782 | 89.1041 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 90.2552 | seriola_lalandi_dorsalis | ENSSLDP00000014993 | 98.2673 | 96.2871 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 90.2552 | stegastes_partitus | ENSSPAP00000003056 | 98.2673 | 96.2871 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.5592 | 88.1671 | anabas_testudineus | ENSATEP00000060444 | 94.1463 | 92.6829 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.3271 | 86.0789 | larimichthys_crocea | ENSLCRP00005024396 | 83.3333 | 80.303 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.7912 | 86.0789 | oryzias_sinensis | ENSOSIP00000047991 | 85.8093 | 82.2616 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.6473 | 89.0951 | oryzias_javanicus | ENSOJAP00000008908 | 97.7723 | 95.0495 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.5592 | 84.9188 | cynoglossus_semilaevis | ENSCSEP00000023220 | 96.0199 | 91.0448 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 90.4872 | amphiprion_percula | ENSAPEP00000013601 | 98.2673 | 96.5347 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 87.007 | nothobranchius_furzeri | ENSNFUP00015010136 | 92.3256 | 87.2093 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.3434 | 90.4872 | pundamilia_nyererei | ENSPNYP00000012899 | 98.5149 | 96.5347 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 56.1485 | 54.9884 | neolamprologus_brichardi | ENSNBRP00000004279 | 99.5885 | 97.5309 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.0951 | 87.239 | mastacembelus_armatus | ENSMAMP00000061811 | 92.0863 | 90.1679 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.3434 | 90.4872 | maylandia_zebra | ENSMZEP00005012937 | 98.5149 | 96.5347 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 79.5824 | 74.0139 | eptatretus_burgeri | ENSEBUP00000015398 | 88.8601 | 82.6425 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.2552 | 83.9907 | latimeria_chalumnae | ENSLACP00000003639 | 95.5774 | 88.9435 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.6473 | 88.1671 | lepisosteus_oculatus | ENSLOCP00000017808 | 96.1071 | 92.4574 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.3271 | 83.5267 | clupea_harengus | ENSCHAP00000009965 | 95.297 | 89.1089 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 87.935 | danio_rerio | ENSDARP00000018923 | 97.7832 | 93.3498 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.9513 | 84.4548 | danio_rerio | ENSDARP00000058912 | 96.5517 | 89.6552 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.4872 | 84.6868 | astyanax_mexicanus | ENSAMXP00000040489 | 96.0591 | 89.9015 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 83.7587 | 80.0464 | astyanax_mexicanus | ENSAMXP00000052586 | 94.5026 | 90.3141 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.3434 | 87.471 | carassius_auratus | ENSCARP00000131018 | 98.0296 | 92.8571 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 41.5313 | 41.0673 | carassius_auratus | ENSCARP00000086275 | 100 | 98.8827 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 87.007 | 81.4385 | carassius_auratus | ENSCARP00000034795 | 91.9118 | 86.0294 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.7193 | 85.3828 | ictalurus_punctatus | ENSIPUP00000007540 | 96.3054 | 90.6404 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.2552 | 85.1508 | electrophorus_electricus | ENSEEEP00000026823 | 89.8383 | 84.7575 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 88.6311 | 83.5267 | electrophorus_electricus | ENSEEEP00000036574 | 92.4939 | 87.1671 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.4153 | 87.703 | esox_lucius | ENSELUP00000062743 | 91.4153 | 87.703 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.5592 | 84.2227 | esox_lucius | ENSELUP00000067467 | 88.1279 | 82.8767 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 87.703 | 84.2227 | oncorhynchus_kisutch | ENSOKIP00005007751 | 93.7965 | 90.0744 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.4153 | 87.239 | oncorhynchus_kisutch | ENSOKIP00005100594 | 69.8582 | 66.6667 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 87.703 | 83.9907 | oncorhynchus_kisutch | ENSOKIP00005022182 | 91.7476 | 87.8641 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.2552 | 86.7749 | oncorhynchus_kisutch | ENSOKIP00005010485 | 94.8781 | 91.2195 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.5754 | 87.471 | oncorhynchus_mykiss | ENSOMYP00000038710 | 93.662 | 88.4977 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.2552 | 86.5429 | oncorhynchus_mykiss | ENSOMYP00000005679 | 94.8781 | 90.9756 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.4153 | 87.703 | oncorhynchus_mykiss | ENSOMYP00000012589 | 91.4153 | 87.703 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.6473 | 87.703 | salmo_salar | ENSSSAP00000034456 | 92.0746 | 88.1119 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 86.7749 | 81.9025 | salmo_salar | ENSSSAP00000057821 | 88.6256 | 83.6493 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.0232 | 86.7749 | salmo_salar | ENSSSAP00000034434 | 95.098 | 91.6667 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.2552 | 86.7749 | salmo_salar | ENSSSAP00000045937 | 94.8781 | 91.2195 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 88.6311 | 84.6868 | salmo_trutta | ENSSTUP00000028421 | 93.3985 | 89.2421 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.0232 | 86.7749 | salmo_trutta | ENSSTUP00000036151 | 95.098 | 91.6667 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 88.8631 | 84.2227 | salmo_trutta | ENSSTUP00000017688 | 91.6268 | 86.8421 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 88.6311 | 83.9907 | salmo_trutta | ENSSTUP00000104035 | 89.2523 | 84.5794 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 87.935 | 83.2947 | gadus_morhua | ENSGMOP00000041226 | 84.4098 | 79.9555 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 41.5313 | 39.9072 | ailuropoda_melanoleuca | ENSAMEP00000040822 | 100 | 96.0894 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.1114 | 87.239 | erpetoichthys_calabaricus | ENSECRP00000016088 | 97.7832 | 92.6108 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.9513 | 86.0789 | pygocentrus_nattereri | ENSPNAP00000037146 | 96.0784 | 90.9314 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.4872 | 84.9188 | pygocentrus_nattereri | ENSPNAP00000020250 | 96.0591 | 90.1478 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.9513 | 87.007 | hucho_hucho | ENSHHUP00000090331 | 86.5342 | 82.7815 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.6473 | 87.471 | hucho_hucho | ENSHHUP00000013021 | 96.577 | 92.176 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.1833 | 87.471 | hucho_hucho | ENSHHUP00000091422 | 96.5602 | 92.629 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 80.2784 | 75.6381 | hucho_hucho | ENSHHUP00000067080 | 90.3394 | 85.1175 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 87.703 | 81.2065 | sinocyclocheilus_grahami | ENSSGRP00000075406 | 85.1351 | 78.8288 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.4153 | 87.703 | oncorhynchus_tshawytscha | ENSOTSP00005081965 | 91.4153 | 87.703 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 87.007 | 83.2947 | oncorhynchus_tshawytscha | ENSOTSP00005044597 | 90.1442 | 86.2981 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 88.3991 | 83.2947 | oncorhynchus_tshawytscha | ENSOTSP00005027816 | 92.2518 | 86.9249 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.2552 | 86.7749 | oncorhynchus_tshawytscha | ENSOTSP00005002334 | 94.8781 | 91.2195 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 92.3434 | 87.935 | paramormyrops_kingsleyae | ENSPKIP00000013940 | 98.0296 | 93.3498 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.8793 | 87.239 | paramormyrops_kingsleyae | ENSPKIP00000014400 | 97.5369 | 92.6108 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 91.4153 | 87.471 | scleropages_formosus | ENSSFOP00015050395 | 91.6279 | 87.6744 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 90.9513 | 84.9188 | scleropages_formosus | ENSSFOP00015035621 | 96.3145 | 89.9263 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.3271 | 85.1508 | denticeps_clupeoides | ENSDCDP00000057388 | 92.5481 | 88.2212 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.0951 | 84.4548 | myripristis_murdjan | ENSMMDP00005043053 | 90.7801 | 86.052 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.5592 | 84.6868 | cyprinus_carpio_carpio | ENSCCRP00000157819 | 93.9173 | 88.8078 |
| ENSBSLG00000001086 | betta_splendens | ENSBSLP00000001920 | 89.7912 | 85.1508 | cyprinus_carpio_carpio | ENSCCRP00000150061 | 98.4733 | 93.3842 |
| Go ID |
Go term |
No. evidence |
Entries |
Species |
Category |