| RBP Type |
N.A. |
| Diseases |
|
| Drug |
|
| Main interacting RNAs |
|
| Moonlighting functions |
|
| Localizations |
|
|
BulkPerturb-seq
|
|
| Ensembl ID | ENSAPEG00000018096 | Gene ID |
|
Accession |
- |
| Symbol |
eef1db |
Alias |
- |
Full Name |
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| Status |
Confidence |
Length |
8230 bases |
Strand |
Plus strand |
| Position |
2 : 29623168 - 29631397 |
RNA binding domain |
EF1_GNE |
| Summary |
N.A. |
RNA binding domains (RBDs)

| Protein |
Domain |
Pfam ID |
E-value |
Domain number |
| ENSAPEP00000025568 |
EF1_GNE |
PF00736 |
8.4e-37 |
1 |
| ENSAPEP00000025531 |
EF1_GNE |
PF00736 |
1e-36 |
1 |
| ENSAPEP00000025625 |
EF1_GNE |
PF00736 |
1.1e-36 |
1 |
| ENSAPEP00000025843 |
EF1_GNE |
PF00736 |
1.8e-36 |
1 |
| ENSAPEP00000025851 |
EF1_GNE |
PF00736 |
3.4e-36 |
1 |
| ENSAPEP00000025810 |
EF1_GNE |
PF00736 |
9.1e-36 |
1 |
| ENSAPEP00000025732 |
EF1_GNE |
PF00736 |
9.7e-36 |
1 |
| ENSAPEP00000025670 |
EF1_GNE |
PF00736 |
1.2e-35 |
1 |
RNA binding proteomes (RBPomes)
| Pubmed ID |
Full Name |
Cell |
Author |
Time |
Doi |
Literatures on RNA binding capacity
| Pubmed ID |
Title |
Author |
Time |
Journal |
| Name |
Transcript ID |
bp |
Protein |
Translation ID |
| - |
ENSAPET00000026204 |
846 |
281aa |
ENSAPEP00000025531 |
| - |
ENSAPET00000026242 |
795 |
264aa |
ENSAPEP00000025568 |
| - |
ENSAPET00000026300 |
852 |
283aa |
ENSAPEP00000025625 |
| - |
ENSAPET00000026345 |
777 |
258aa |
ENSAPEP00000025670 |
| - |
ENSAPET00000026411 |
693 |
230aa |
ENSAPEP00000025732 |
| - |
ENSAPET00000026489 |
672 |
223aa |
ENSAPEP00000025810 |
| - |
ENSAPET00000026523 |
366 |
121aa |
ENSAPEP00000025843 |
| - |
ENSAPET00000026531 |
450 |
149aa |
ENSAPEP00000025851 |
| Pathway ID |
Pathway Name |
Source |
| | |
| ensgID |
Trait |
pValue |
Pubmed ID |
| ensgID | SNP | Chromosome | Position |
Trait | PubmedID | Or or BEAT |
EFO ID |
|---|
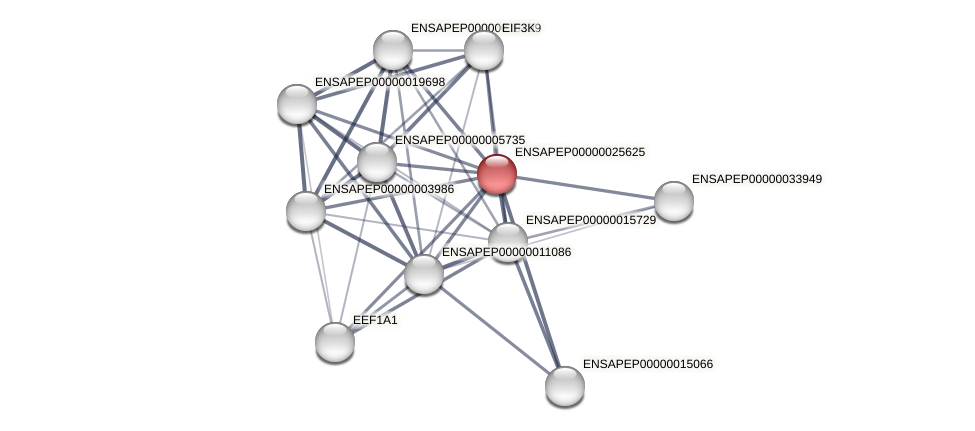
Protein-Protein Interaction (PPI)
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000034714 | 43.0769 | 38.4615 | amphiprion_percula | ENSAPEP00000025625 | 69.2579 | 61.8375 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000021908 | 20.712 | 10.356 | amphiprion_percula | ENSAPEP00000025625 | 45.2297 | 22.6148 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000011086 | 63.7209 | 54.4186 | amphiprion_percula | ENSAPEP00000025625 | 48.4099 | 41.3428 |
| Ensembl ID |
Source |
Target |
| Species |
Protein ID |
Perc_pos |
Perc_id |
Species |
Protein ID |
Perc_pos |
Perc_id |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 86.5724 | 82.3322 | fundulus_heteroclitus | ENSFHEP00000006536 | 34.9501 | 33.2382 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 87.2792 | 80.5654 | poecilia_reticulata | ENSPREP00000015199 | 30.4562 | 28.1134 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 90.106 | 84.0989 | xiphophorus_maculatus | ENSXMAP00000021630 | 35.3186 | 32.964 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 89.3993 | 81.9788 | oryzias_latipes | ENSORLP00000015598 | 35.5337 | 32.5843 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 84.8057 | 79.1519 | cyclopterus_lumpus | ENSCLMP00005035288 | 89.2193 | 83.2714 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 86.5724 | 81.2721 | haplochromis_burtoni | ENSHBUP00000020130 | 34.0751 | 31.9889 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 90.106 | 84.4523 | astatotilapia_calliptera | ENSACLP00000036589 | 35.3186 | 33.1025 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 87.2792 | 81.6254 | oreochromis_niloticus | ENSONIP00000013836 | 34.022 | 31.8182 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 91.8728 | 86.9258 | sparus_aurata | ENSSAUP00010029054 | 36.2117 | 34.2618 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 88.3392 | 83.0389 | lates_calcarifer | ENSLCAP00010047977 | 31.0559 | 29.1925 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 91.5194 | 87.9859 | dicentrarchus_labrax | ENSDLAP00005036088 | 35.9722 | 34.5833 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 85.8657 | 80.9187 | cyprinodon_variegatus | ENSCVAP00000003918 | 33.5172 | 31.5862 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 84.4523 | 79.1519 | kryptolebias_marmoratus | ENSKMAP00000015738 | 84.7518 | 79.4326 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 86.5724 | 79.1519 | oryzias_melastigma | ENSOMEP00000012208 | 83.9041 | 76.7123 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 87.9859 | 80.9187 | takifugu_rubripes | ENSTRUP00000071826 | 36.0347 | 33.1404 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 90.4594 | 84.4523 | neolamprologus_brichardi | ENSNBRP00000029429 | 85.6187 | 79.9331 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 91.5194 | 87.2792 | seriola_dumerili | ENSSDUP00000029930 | 35.823 | 34.1632 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 90.4594 | 85.5124 | poecilia_formosa | ENSPFOP00000000267 | 32.9049 | 31.1054 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 88.3392 | 83.7456 | poecilia_latipinna | ENSPLAP00000003717 | 89.9281 | 85.2518 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 87.6325 | 84.0989 | cottoperca_gobio | ENSCGOP00000016769 | 34.4444 | 33.0556 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 86.2191 | 80.5654 | scophthalmus_maximus | ENSSMAP00000040892 | 33.9833 | 31.7549 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 88.6926 | 80.5654 | labrus_bergylta | ENSLBEP00000005898 | 21.6007 | 19.6213 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 91.8728 | 90.4594 | acanthochromis_polyacanthus | ENSAPOP00000034283 | 35.9613 | 35.408 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 94.3463 | 93.6396 | amphiprion_ocellaris | ENSAOCP00000026137 | 36.8276 | 36.5517 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 84.8057 | 78.4452 | gasterosteus_aculeatus | ENSGACP00000007479 | 41.7391 | 38.6087 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 73.4982 | 68.5512 | gasterosteus_aculeatus | ENSGACP00000001840 | 89.2704 | 83.2618 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 86.2191 | 80.5654 | tetraodon_nigroviridis | ENSTNIP00000011062 | 46.0377 | 43.0189 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 75.6184 | 70.318 | nothobranchius_furzeri | ENSNFUP00015019762 | 82.9457 | 77.1318 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 90.106 | 84.4523 | maylandia_zebra | ENSMZEP00005022406 | 35.3186 | 33.1025 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 85.159 | 78.0919 | oryzias_javanicus | ENSOJAP00000013652 | 88.9299 | 81.5498 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 76.6784 | 72.4382 | mastacembelus_armatus | ENSMAMP00000020142 | 82.5095 | 77.9468 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 86.5724 | 81.2721 | amphilophus_citrinellus | ENSACIP00000013942 | 33.4699 | 31.4208 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 81.9788 | 75.9717 | hippocampus_comes | ENSHCOP00000007144 | 72.7273 | 67.3981 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 90.106 | 84.4523 | pundamilia_nyererei | ENSPNYP00000030150 | 85.5705 | 80.2013 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 90.8127 | 85.159 | anabas_testudineus | ENSATEP00000002274 | 87.415 | 81.9728 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 87.9859 | 82.6855 | larimichthys_crocea | ENSLCRP00005028681 | 80.8442 | 75.974 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 52.2968 | 44.8763 | larimichthys_crocea | ENSLCRP00005028804 | 67.8899 | 58.2569 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 66.7845 | 57.5972 | cynoglossus_semilaevis | ENSCSEP00000003748 | 45.6522 | 39.372 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 49.1166 | 44.523 | cynoglossus_semilaevis | ENSCSEP00000013247 | 87.9747 | 79.7468 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 90.8127 | 86.2191 | seriola_lalandi_dorsalis | ENSSLDP00000022298 | 87.415 | 82.9932 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 91.5194 | 87.2792 | sander_lucioperca | ENSSLUP00000022813 | 88.3959 | 84.3003 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 80.212 | 76.3251 | betta_splendens | ENSBSLP00000027914 | 92.6531 | 88.1633 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 93.6396 | 90.4594 | stegastes_partitus | ENSSPAP00000022554 | 88.9262 | 85.906 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 83.3922 | 76.6784 | oryzias_sinensis | ENSOSIP00000008329 | 81.0997 | 74.5704 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 44.8763 | 32.8622 | caenorhabditis_elegans | F54H12.6.1 | 59.6244 | 43.662 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 49.47 | 37.4558 | drosophila_melanogaster | FBpp0311691 | 63.0631 | 47.7477 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 59.0106 | 44.523 | drosophila_melanogaster | FBpp0079542 | 65.2344 | 49.2188 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 49.47 | 34.9823 | ciona_intestinalis | ENSCINP00000023206 | 62.2222 | 44 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 67.1378 | 55.1237 | petromyzon_marinus | ENSPMAP00000008388 | 75.6972 | 62.1514 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 82.6855 | 71.3781 | lepisosteus_oculatus | ENSLOCP00000010373 | 34.6154 | 29.8817 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 86.9258 | 75.6184 | esox_lucius | ENSELUP00000000469 | 33.6986 | 29.3151 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 80.5654 | 70.6714 | oncorhynchus_kisutch | ENSOKIP00005056892 | 28.6792 | 25.1572 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 83.3922 | 71.7314 | salmo_salar | ENSSSAP00000031655 | 30.7692 | 26.4668 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 84.8057 | 73.4982 | salmo_salar | ENSSSAP00000014266 | 32.3015 | 27.9946 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 84.8057 | 74.2049 | oncorhynchus_mykiss | ENSOMYP00000070292 | 31.9149 | 27.9255 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 84.4523 | 73.4982 | salmo_trutta | ENSSTUP00000026992 | 33.2869 | 28.9694 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 84.8057 | 74.5583 | salmo_trutta | ENSSTUP00000003020 | 32.0856 | 28.2086 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 83.0389 | 72.4382 | gadus_morhua | ENSGMOP00000015042 | 86.3971 | 75.3676 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 8.12721 | 3.18021 | ochotona_princeps | ENSOPRP00000015286 | 4.29104 | 1.6791 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 61.4841 | 44.8763 | ciona_savignyi | ENSCSAVP00000004915 | 65.9091 | 48.1061 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 50.53 | 37.8092 | ciona_savignyi | ENSCSAVP00000018739 | 63.8393 | 47.7679 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 84.0989 | 73.4982 | hucho_hucho | ENSHHUP00000014422 | 33.6634 | 29.4201 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 71.7314 | 63.6042 | hucho_hucho | ENSHHUP00000017964 | 82.1862 | 72.8745 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 85.159 | 74.5583 | oncorhynchus_tshawytscha | ENSOTSP00005086665 | 32.0905 | 28.0959 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 85.159 | 74.5583 | oncorhynchus_tshawytscha | ENSOTSP00005104234 | 32.0905 | 28.0959 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 82.6855 | 71.3781 | scleropages_formosus | ENSSFOP00015010390 | 37.6812 | 32.5282 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 80.5654 | 70.318 | paramormyrops_kingsleyae | ENSPKIP00000015673 | 37.4384 | 32.6765 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 39.576 | 33.5689 | denticeps_clupeoides | ENSDCDP00000017001 | 89.6 | 76 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 39.576 | 33.5689 | denticeps_clupeoides | ENSDCDP00000017397 | 89.6 | 76 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 88.3392 | 81.2721 | myripristis_murdjan | ENSMMDP00005031006 | 81.4332 | 74.9186 |
| ENSAPEG00000018096 | amphiprion_percula | ENSAPEP00000025625 | 42.0495 | 30.3887 | saccharomyces_cerevisiae | YAL003W | 57.767 | 41.7476 |
| Go ID |
Go term |
No. evidence |
Entries |
Species |
Category |